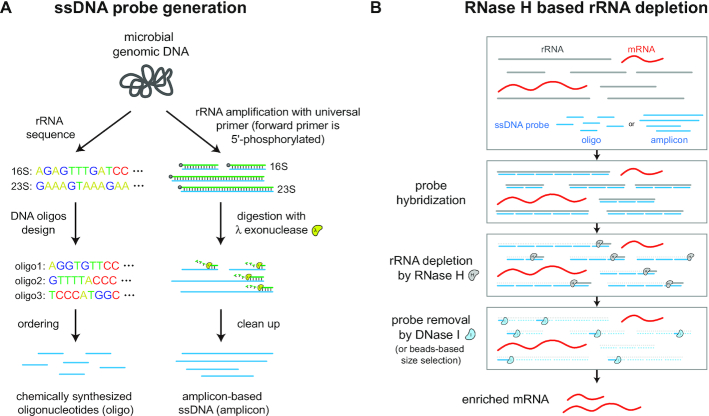
Figure 1.
Workflow for bacterial RNase H based rRNA depletion. (A) Probes used for depletion can be either designed and chemically synthesized from known rRNA sequences (oligo-based) or generated by PCR from genomic DNA with 5′-phosphorylated forward primers and subsequent lambda exonuclease digestion (amplicon-based). (B) Probes are then hybridized to total RNA and the rRNA bound by the ssDNA probes is degraded by RNase H. At last, all remaining probes are degraded by DNase I or removed by SPRI beads-based size selection, resulting in enriched mRNAs.