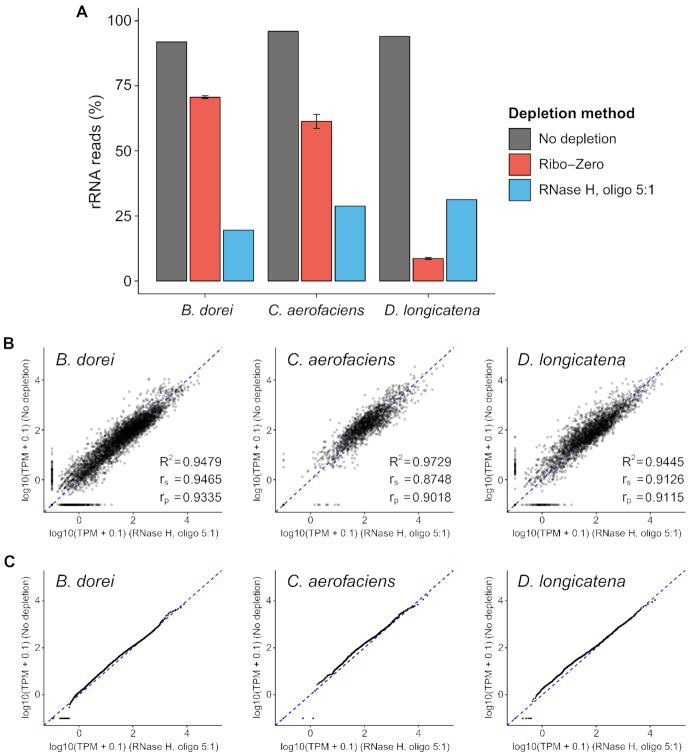
Figure 3.
Application of RNase H based rRNA depletion with oligos to three diverse gut microbiota species. (A) Proportion of rRNA-aligning reads of total mapped reads (% rRNA reads), for un-depleted, Ribo-Zero depleted and RNase H depleted samples under optimized reaction condition across three microbiota species from different phyla. The same RNA samples used for Ribo-Zero depletion were pooled together to yield the RNA used for RNase H reaction. (B andC) Consistency in transcriptome between rRNA-depleted samples and un-depleted samples in terms of expression correlation (B) and expression distribution (C). TPM indicates transcripts per million for each CDS. (C) Points lie on a straight line in Quantile-Quantile (Q-Q) plots if there is no global shift in the distribution of expression profile between depleted samples and un-depleted samples.