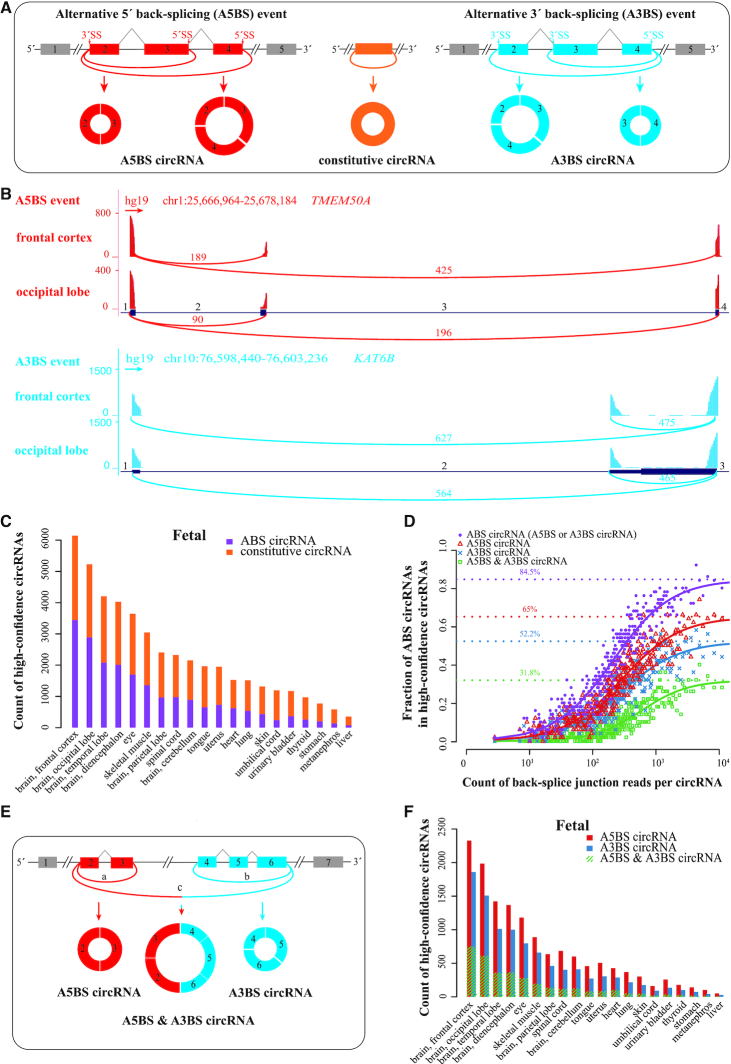
Figure 1.
ABS events of circRNAs are prevalent in human tissue transcriptomes. (A) Schematic diagrams of two types of ABS events and three types of circRNAs. Colored boxes, exons. Black lines, introns. Gray polylines, canonical splicing. Colored arcs, back-splicing. Circles, circRNAs. (B) An example A5BS event (top panel) and an example A3BS event (bottom panel) in genes TMEM50A and KAT6B in human frontal cortex and occipital lobe tissues, respectively. The number of back-splice junction reads for each circRNA is displayed as red text above the corresponding arc. The indices of circRNA-forming introns are labeled in black text above the introns: introns 1, 2, 3 and 4 for TMEM50A and introns 1, 2 and 3 for KAT6B. Red and blue arcs, back-splicing. (C) The count of ABS and constitutive circRNAs among high-confidence circRNAs (≥ 0.1 RPM) for human fetal tissues. (D) Mean fraction of ABS circRNAs in groups of 50 circRNAs, binned by their pooled back-splice junction reads. The fraction of ABS circRNAs at sufficient sequencing depth was estimated to be the asymptote of the best-fit sigmoid curve (purple for ABS circRNAs, red for A5BS circRNAs, blue for A3BS circRNAs, and green for A5BS & A3BS circRNAs). Points show the fraction of ABS circRNAs in each bin. (E) Schematic diagrams of an example A5BS & A3BS circRNA. Colored boxes, exons. Black lines, introns. Colored arcs, back-splicing. Circles, circRNAs. (F) The count of A5BS & A3BS circRNAs as special cases of A5BS circRNAs and A3BS circRNAs for human fetal tissues.