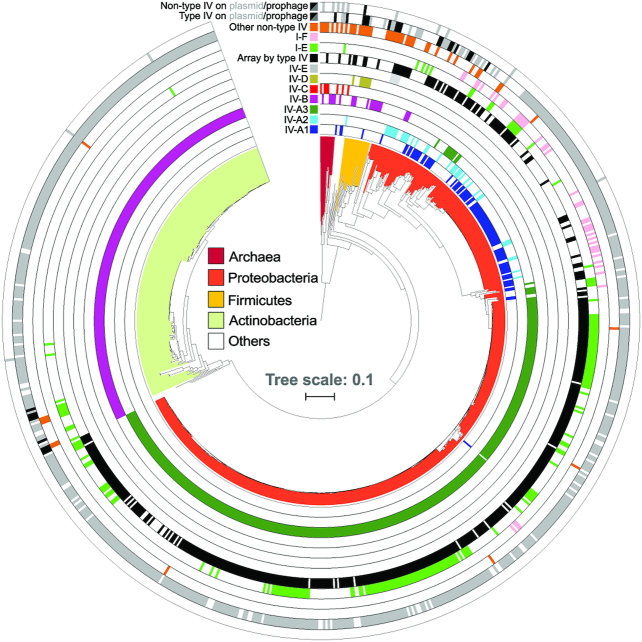
Figure 2.
Distribution of type IV loci across prokaryotic taxa and MGE types. Phylogenetic tree based on 16S rRNA gene sequences of all bacteria and archaea that carry type IV CRISPR–Cas systems. Concentric rings denote the presence or absence of type IV and other co-occurring non-type IV CRISPR–Cas loci in the same genomes, colour-coded according to the subtype/variant to which they belong. All non-type IV systems, except I-E (light green) and I-F (pink), were merged into one lane (orange) for visualization purposes. Type IV effector cas operons for which an associated CRISPR array was detected are shown (black). Based on genomic context analyses (Methods), CRISPR–Cas systems predicted to be encoded by plasmid-like elements (grey) or (pro)phages/viruses (black) are shown (Supplementary Data S2), for both type IV and non-type IV loci (two outermost ring lanes). 758 (of 883) identified type IV loci are displayed on the tree; for the remainder no 16S rRNA gene sequence was found in the genome.