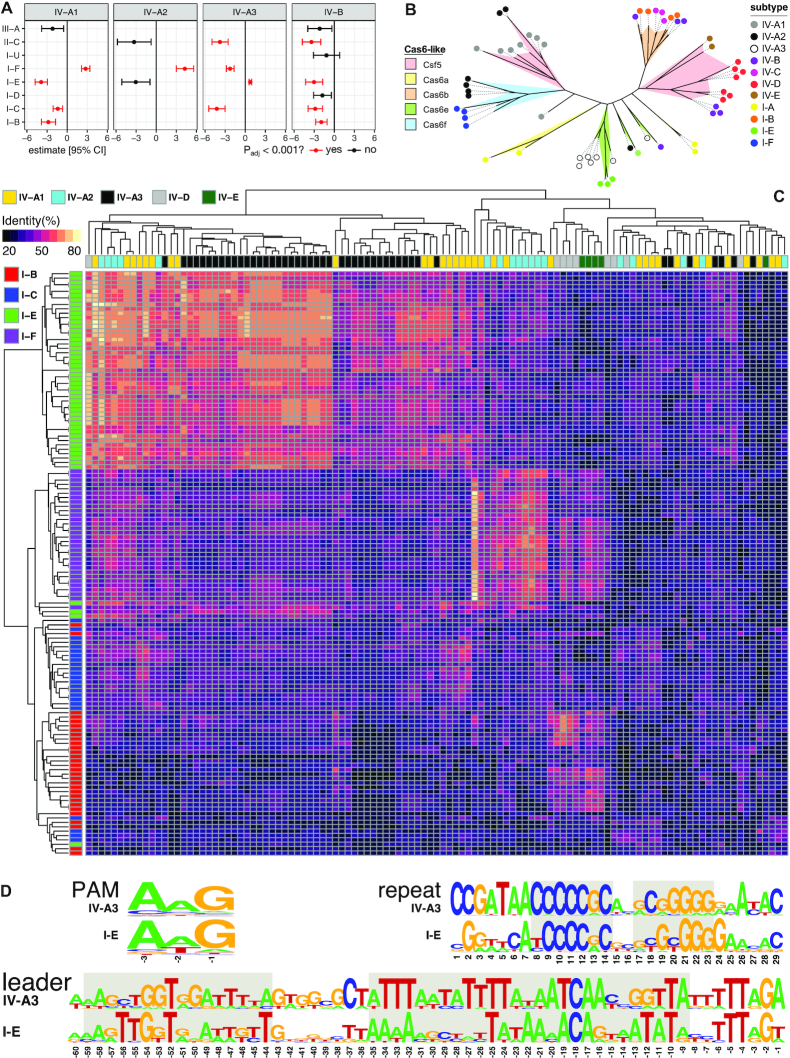
Figure 4.
Interactions between type IV CRISPR–Cas systems and other co-encoded CRISPR–Cas systems in a host. (A) Co-occurrence analysis between type IV and non-type IV systems. Estimates are from phylogenetic logistic regressions, with P-values fdr-adjusted. Only estimates with standard errors <10 are shown. (B) Unrooted phylogenetic tree for Cas6/Csf3 built with representatives covering the diversity of type IV and type I subtypes/variants detected in this study. Each cluster is coloured according to the cas6-like family it corresponds to, and the coloured dot at the end of branches indicates the specific RISPR–Cas subtype/variant encoding such a Cas6-like protein. (C) Heat map depicting CRISPR repeat similarity of co-occurring CRISPR–Cas subtypes/variants clustered by average linkage hierarchical clustering. (D) PAM, consensus CRISPR repeat and leader sequence logos for the positively correlated subtypes IV-A3 and I-E. The short semi-palindromic repeats at the centre of the consensus repeat that are used as anchor sequences by the Cas1–Cas2e complex are highlighted in grey, as well as the conserved leader sequences comprising the binding sites for the Cas1–Cas2e complex (left) and the IHF (right).