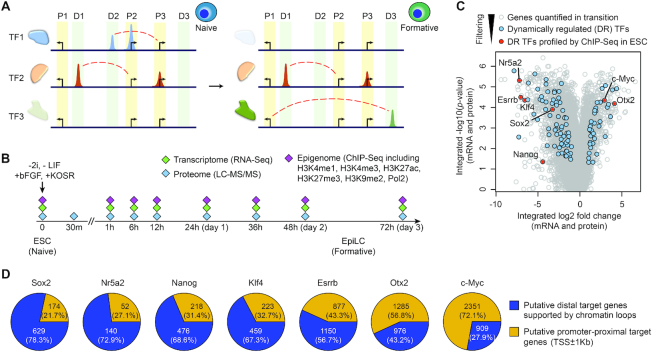
Figure 1.
Identification of dynamically regulated TFs in pluripotency progression. (A) Schematics of TF expression and their binding at promoter-proximal (P1, P2 and P3) and distal (D1, D2 and D3) sites during pluripotency progression from naive to formative states. Dash lines (red) represent interactions between distal TF binding sites and their target genes. (B) Schematic summary of the time-course trans-omic dataset utilised in this study for reconstructing and characterising the transcriptional networks in pluripotency progression from naive ESCs to EpiLCs that represent formative state. (C) Volcano plot of genes profiled on both transcriptome and proteome levels in the trans-omic dataset (27). TFs that are dynamically regulated (DR) during the ESC to EpiLC transition are highlighted in blue and within these DR TFs, those that have been profiled previously using ChIP-seq in ESCs are highlighted in red. (D) Pie charts showing the distribution of promoter-proximal target genes (TSS ± 1 kb) and putative distal target genes (TSS > 1 kb) with chromatin loops (Pol2-ChIA-PET) for each TF according to its ChIP-seq profile in ESCs.