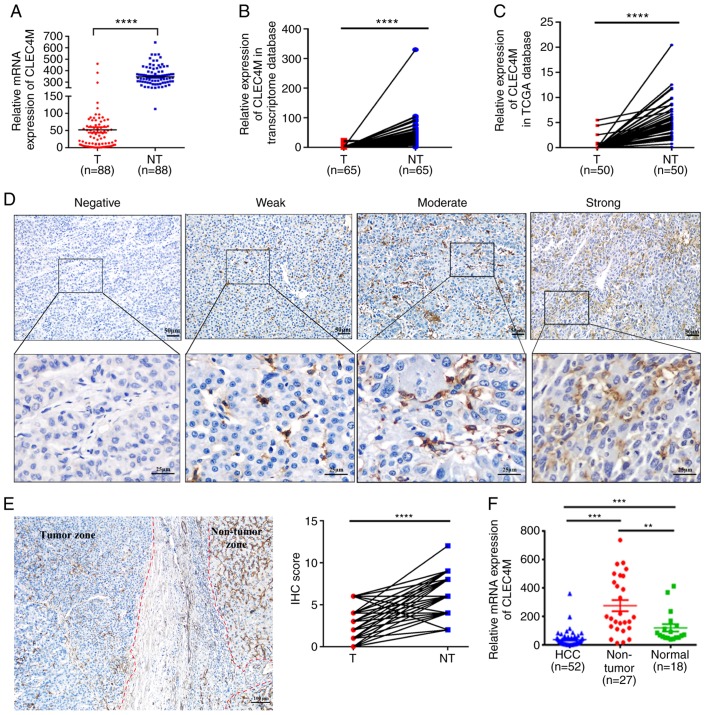
Figure 1.
Determination of CLEC4M expression level in HCC tumor vs. adjacent non-tumor tissues. (A) RT-qPCR results of CLEC4M mRNA expression in 88 pairs of HCC tumor samples and their corresponding adjacent non-tumor tissues; paired Student's t-test. (B) transcriptomic analysis of the relative expression of CLEC4M in HCC tissues vs. adjacent non-tumor tissues. N=65, paired Student's t-test. (C) Analysis of CLEC4M mRNA expression in The Cancer Genome Atlas database; n=50; paired t-test. (D) CLEC4M expression levels in HCC tumor tissues. CLEC4M expression was semi-quantitatively divided into four groups: negative staining, weak-positive staining, moderate-positive staining and strong-positive staining (magnification, ×40 and ×200); (E) representative immunohistochemical section of HCC border indicating the decreased CLEC4M expression in the tumor tissue (magnification, ×200; (F) RT-qPCR results of CLEC4M mRNA expression in HCC (n=52), non-tumor tissues (n=27) and normal hepatic tissues (n=18); One-way ANOVA combined with Tukey's multiple comparisons test (post-hoc) was applied to perform the comparisons between groups. **P<0.01, ***P<0.001 and ****P<0.0001. T, tumor tissue; NT, non-tumor tissue. HCC, hepatocellular carcinoma; CLEC4M, C-type lectin domain family 4 member M; RFS, recurrence free survival; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IHC, immunohistochemical.