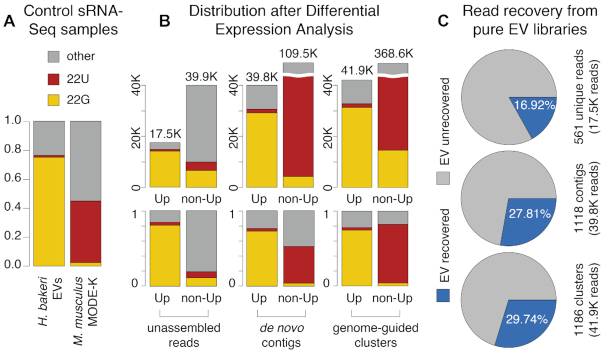
Figure 4.
Evaluation of differential expression results. Reads were categorized as 22G (yellow), 22U (red), or other (grey) based on length and first-nucleotide. (A) sRNA profiles of control samples: purified H. bakeri Extracellular Vesicles (EVs) and untreated MODE-K cells. Bar height represents the fraction of all reads. (B) sRNA profiles of unassembled reads, de novo contigs and genome-guided clusters. For each of these sets, there are two bars, the first one represents differentially expressed up-regulated elements (Up) and the second, elements that lack evidence for differential expression in this direction (non-Up). Bar height represents the number of reads (top) or the fraction of reads (bottom) belonging to these categories. (C) Percent of reads from pure H. bakeri EV libraries, recovered during differential expression analysis of MODE-K cells treated with EVs. Each circle represents the total reads from EV libraries. The recovered fractions are indicated in blue. The numbers of H. bakeri differentially expressed elements are shown to the right, as well as total read counts (from B).