FIGURE 5.
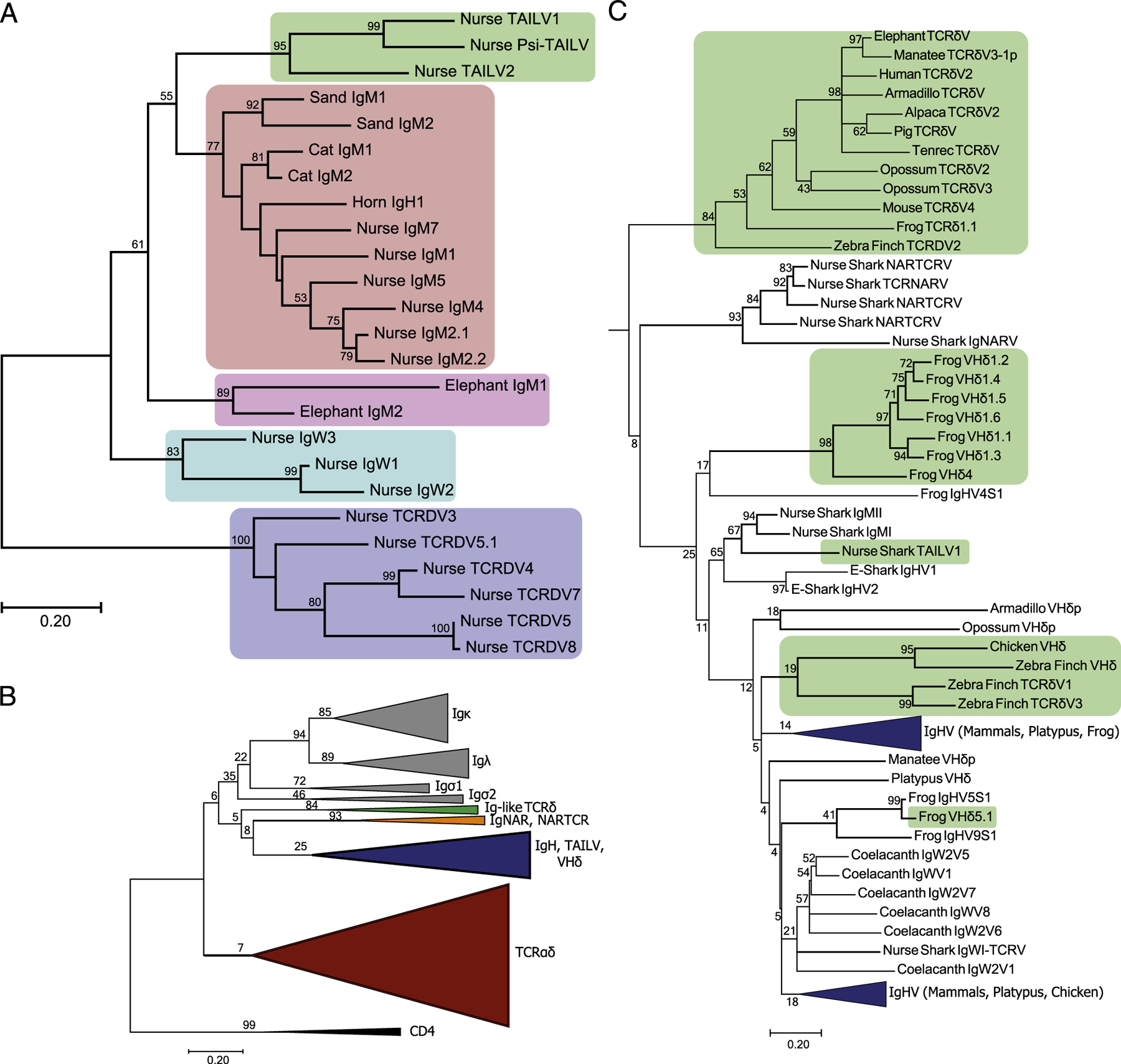
Phylogenetic analysis of Ag receptor V segments unveils a novel lineage of TCRδ-associated IgH-like V segments (TAILV) in cartilaginous fish. (A) An amino acid alignment of chondrichthyes V segments, trimmed to the start of FR1 and ended at the conserved Cys of FR3, was used to construct a maximum likelihood tree. Species included in the alignment were Ginglymostoma cirratum (nurse), Scyliorhinus canicula (cat), Heterodontus francisci (horn), Carcharhinus plumbeus (sand), and Callorhinchus milii (elephant). The percentage of trees supporting branches after 1000 Bootstrap replications is displayed for bifurcations with >50%. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The shaded regions indicate different V segment lineages found in Chondrichthyes, including elasmobranch TAILVs (green), elasmobranch IgMV (red), holocephali IgMV (pink), elasmobranch IgW (teal), and elasmobranch TCRδV (blue). (B) Topology of the jawed vertebrate Ag receptor tree. V segment lineage indicated by color for IgL (gray), Ig-like TCRδV (green), IgNARV/NARTCRV (orange), IgH (blue, includes TAILV and VHδ), and TCRαδV (red). The tree includes representative sequences from human, mouse, pig, tenrec, elephant, alpaca, manatee, opossum, platypus, zebra finch, chicken, frog, coelacanth, nurse shark, and elephant shark. (C) Subtree of the IgH and Ig-like V segments. Green shading indicates lineages of Ig-like V segments that are exclusively found on TCR rearrangements, highlighting multiple exchanges of Ig V segments into TCR loci throughout vertebrate evolution.
