Abstract
Melanoma is the most aggressive and lethal type of skin cancer. The aim of the present study was to illustrate the molecular mechanism of makorin ring finger protein 2 (MKRN2) control of melanoma cell proliferation. The expression level of MKRN2 was detected in human malignant melanoma cell lines by immunoblotting and reverse transcription-quantitative PCR. Short hairpin RNAs for MKRN2 were designed and transfected into melanoma cells, and the proliferation of these cells was detected by MTT and colony formation assays. The interaction of MKRN2 with P53 was detected by co-immunoprecipitation and glutathione S-transferase pulldown assays. The ubiquitination of P53 by MKRN2 was detected by in vitro ubiquitination assays. A P53-knockout cell line was generated using the CRISPR-Cas9 method. MKRN2 exhibited higher expression levels in melanoma cells, and downregulation of MKRN2 inhibited melanoma cell growth in a P53-dependent manner. MKRN2 regulated melanoma cell proliferation by interacting and ubiquitylating P53, which suggests that MKRN2 may be a potential therapeutic target for melanoma.
Keywords: melanoma, makorin ring finger protein 2, P53, proliferation
Introduction
Melanoma is the most aggressive and lethal type of skin cancer worldwide (1). Although it accounts for only 4% of all cases of skin cancer, it causes the highest number of cases of skin cancer-associated mortality worldwide due to its high metastasis (2,3). Following lung and breast cancer, melanoma is the third most common reason for brain metastases (4). Melanoma spreads to the brain in ≤75% of patients with melanoma (5). Brain metastases cause mortality in 95% of patients with high class (Class III) (6). The incidence of primary melanoma has been increasing rapidly and doubles every 10–14 years, from 2006 to 2013 (7). Different anti-melanoma strategies have been developed and used in clinical treatment, including chemotherapy, immunotherapy and targeted therapy (8). Targeted therapy is widely used in cancer treatment due to its small side effects compared with other strategies (9). BRAF inhibitors, Sorafenib, Vemurafenib, Dabrafenib, mitogen-activated protein kinase kinase inhibitor and Trametinib have been widely used for melanoma treatment, but different inhibitors specifically target particular mechanisms of melanoma occurrence and progression, and one inhibitor usually only treats a small percentage of patients with melanoma (2,10). Therefore, novel targets for melanoma treatment are urgently required.
Makorin ring finger protein 2 (MKRN2) belongs to the makorin RING zinc finger family that encodes putative ribonucleoproteins with a distinctive array of zinc finger domains (11,12). Makorins are zinc finger proteins with a typical C3HC4 motif (the RING domain), associated with arrays of one to four C3H domains, and represent a type of zinc finger found in a variety of ribonucleoproteins (13,14). MKRN2 harbours four C3H zinc fingers and a signature C3HC4 RING zinc finger domain (15). The RING domain is responsible for ubiquitin ligase activity, leading to monoubiquitination and/or the synthesis of polyubiquitin chains on lysine residues (16). To date, two substrates for MKRN2 have been reported on, the p65 subunit of NF-κB (P65) and the p85α subunit of PI3K (PI3Kp85α) (12,17). According to previous studies, MKRN2 is a novel ubiquitin E3 ligase targeting the p65 subunit of NF-κB to negatively regulate inflammatory responses (17), and MKRN2 inhibits the migration and invasion of non-small-cell lung cancer by negatively regulating the PI3K/Akt signalling pathway (12). In the present study, MKRN2 expression was higher in human malignant melanoma cell lines compared with that in normal skin cell lines. Downregulation of MKRN2 inhibited melanoma cell growth accompanying P53 upregulation, and further experiments demonstrated that MKRN2 interacted with and ubiquitylated P53. The present study suggests that MKRN2 may act as a potential therapeutic target for melanoma.
Materials and methods
Plasmid construction
A scramble short hairpin RNA (shRNA) and three MKRN2-targeting shRNAs inserted into pLKO.1 plasmids were purchased from Sigma-Aldrich; Merck KGaA; their sequences are provided in Table I. Single guide RNA (sgRNA) targeting TP53 was designed using an online tool (http://crispr.mit.edu/), as previously described (18). The designed sgRNAs (Table I) were synthesized as oligos (Sangon Biotech Co., Ltd.), annealed and inserted into a PX330 vector that was digested with BbsI. The proteins E1 [ubiquitin like modifier activating enzyme 1 (UBA1)-hexahistidine (His6)], E2 [ubiquitin conjugating enzyme E2 D1 (UBCH5A)-His6] and His6-Ub, and the plasmids pET28a-TP53-His6 and pGEX4T-1-Gst-MKRN2 were provided by Professor Ronggui Hu (Chinese Academy of Sciences, Shanghai, China).
Table I.
Sequences of shRNAs for MKRN2 and sgRNA for TP53.
| shRNA/sgRNA | Target site sequence (5′-3′) |
|---|---|
| MKRN2 | |
| Scramble | GCGCGATAGCGCTAATAATTT |
| shRNA1 | CCTATGGAACTCGGTGCAGAT |
| shRNA2 | GACCTCTTCATGCACCTTTCT |
| shRNA3 | GTCCAGAATGCCGTGTGATAT |
| TP53 | |
| sgRNA | CCATTGTTCAATATCGTCCGGGG |
MKRN2, makorin ring finger protein 2; sgRNA, single guide RNA; shRNA, short hairpin RNA.
Cell culture and transfection
Human malignant melanoma cell lines A375, SK-MEL-28 and WM-115 were purchased from the American Type Culture Collection (ATCC). The immortal human keratinocyte cell line HaCaT was originally purchased from the ATCC, and the primary-cultured normal human epithelial melanocyte cell line NHEM was originally purchased from PromoCell GmbH. These two cell lines were provided by Professor Ronggui Hu (Chinese Academy of Sciences, Shanghai, China). All cell lines were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere of 5% CO2. The plasmids containing MKRN2 shRNAs or TP53 sgRNA (both 3 µg) were transfected into melanoma cells using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and screened by puromycin (Thermo Fisher Scientific, Inc.) for two weeks before subsequent experimentation.
Cell proliferation assay
Cells (3,000 cells/well) stably transfected with the MKRN2 shRNAs were seeded into a 96-well plate. The 0 h time-point was defined as 6 h after the cells were seeded. After 0, 24 or 48 h, the cells were incubated with MTT solution (cat. no. C0009; Beyotime Institute of Biotechnology) for 4 h at 37°C. Subsequently, the product (formazan) was dissolved in DMSO and quantified spectrophotometrically at a wavelength of 570 nm using a microplate reader (Bio-Rad Laboratories, Inc.). Experiments were conducted with six replicates and repeated three times.
Colony formation assay
Cells (3,000 cells/well) stably transfected with the MKRN2 shRNAs were seeded into a 6-well plate. After 7 days, plates were fixed with 4% paraformaldehyde (Merck KGaA) at room temperature for 30 min, stained with 0.1% crystal violet (C0121; Beyotime Institute of Biotechnology), at room temperature for 30 min and washed three times with PBS buffer. Images were obtained using a camera (DSC-W800; Sony Corporation), the number of colonies was counted, and the average number was calculated.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells (100,000 cells/well) using a total RNA kit (cat. no. DP419; Tiangen Biotech Co., Ltd.). cDNA was synthesized using ReverTra Ace qPCR RT Master mix (Toyobo Life Science) at 37°C for 15 min, and 95°C for 5 min, according to the manufacturer's protocol. qPCR was performed on an ABI 7500 fast real-time PCR system using the following conditions: Initial denaturation at 95°C for 2 min; denaturation at 95°C for 5 sec, annealing at 60°C for 20 sec, and elongation 72°C for 34 sec, for 40 cycles (Applied Biosystems; Thermo Fisher Scientific, Inc.) to assess the relative abundances of TP53 and cyclin dependent kinase inhibitor 1A (p21) mRNAs using specific primers (Table II) with staining by SYBR Green (Toyobo Life Science). The relative abundances of TP53 and p21 were normalized to that of the GAPDH gene, using the 2−ΔΔCq method (19,20). All data were obtained from three independent experiments.
Table II.
Sequences of the primers used in reverse transcription-quantitative PCR.
| Target gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| GAPDH | GAGTCAACGGATTTGGTCGTATTG | ATTTGCCATGGGTGGAATCATATTG |
| TP53 | CAGACCTATGGAAACTACTTCCTGA | CTTCATCTGGACCTGGGTCTTC |
| p21 | CTGTCACTGTCTTGTACCCTTGT | GGAGTGGTAGAAATCTGTCATGCT |
p21, cyclin dependent kinase inhibitor 1A.
Co-immunoprecipitation (COIP), immunoprecipitation and immunoblotting
For COIP, A375 cells (500,000 cells/well) were lysed in 500 µl COIP buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA and 1% NP-40, pH 7.6) supplemented with a protease inhibitor cocktail (Roche Diagnostics). Subsequently, the cell lysates were centrifuged at 4°C at 12,000 × g for 10 min, incubated with P53 antibody (1:100; cat. no. 10442-1-AP; ProteinTech Group, Inc.) and Protein G agarose beads (Merck KGaA) overnight at 4°C, then washed three times with COIP buffer. For immunoprecipitation, cells (500,000 cells/well) were lysed in 500 µl immunoprecipitation buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.1% SDS and 1% NP-40, pH 7.6) supplemented with a protease inhibitor cocktail. Subsequently, the cell lysates were centrifuged at 4°C at 12,000 × g for 10 min, incubated with P53 antibody and Protein G agarose beads overnight at 4°C, then washed three times with COIP buffer. The immunoprecipitates were enriched and denatured at 100°C for 10 min in 2X SDS-PAGE loading buffer (50 µl). The inputs, immunoprecipitates and other cell lysates (10 µl/lane) were then subjected to 10% SDS-PAGE, transferred to a PVDF membrane (Bio-Rad Laboratories, Inc.) at 200 mA for 3 h. Following which, the membrane was blocked with 5% skimmed milk at room temperature for 1 h, then incubated with the appropriate antibodies against MKRN2 (1:500 dilution; cat. no. 12238-1-AP; ProteinTech Group, Inc.), P53 (1:2,000 dilution; as aforementioned), P21 (1:1,000 dilution; cat. no. 10355-1-AP; ProteinTech Group, Inc.), MDM2 proto-oncogene (MDM2; 1:500 dilution; cat. no. 19058-1-AP; ProteinTech Group, Inc.), P65 (1:1,000 dilution; cat. no. 10745-1-AP; ProteinTech Group, Inc.), ubiquitin (1:1,000 dilution; sc-47721; Santa Cruz Biotechnology, Inc.) and GAPDH (1:5,000 dilution; cat. no. 60004-1-Ig; ProteinTech Group, Inc.) overnight at 4°C, Subsequently the membranes were washed three times with TBST (50 mM Tris-HCl, 150 mM NaCl and 0.1% Tween-20, pH 7.6) then incubated with secondary antibodies [horseradish peroxidase (HRP)-conjugated Affinipure goat anti-mouse IgG (H+L); cat. no. SA00001-1; 1:5,000 dilution and HRP-conjugated Affinipure goat anti-rabbit IgG (H+L); cat. no. SA00001-2; 1:5,000 dilution; ProteinTech Group, Inc.] at room temperature for 1 h, and washed three times with TBST. The signals were visualized using high-signal ECL western blotting substrate (cat. no. 180-5001) and a Tanon 5200 Imaging system (both Tanon Science and Technology Co., Ltd.). The density of the protein bands in Fig. 1 was calculated using ImageJ software v1.8.0 (National Institutes of Health).
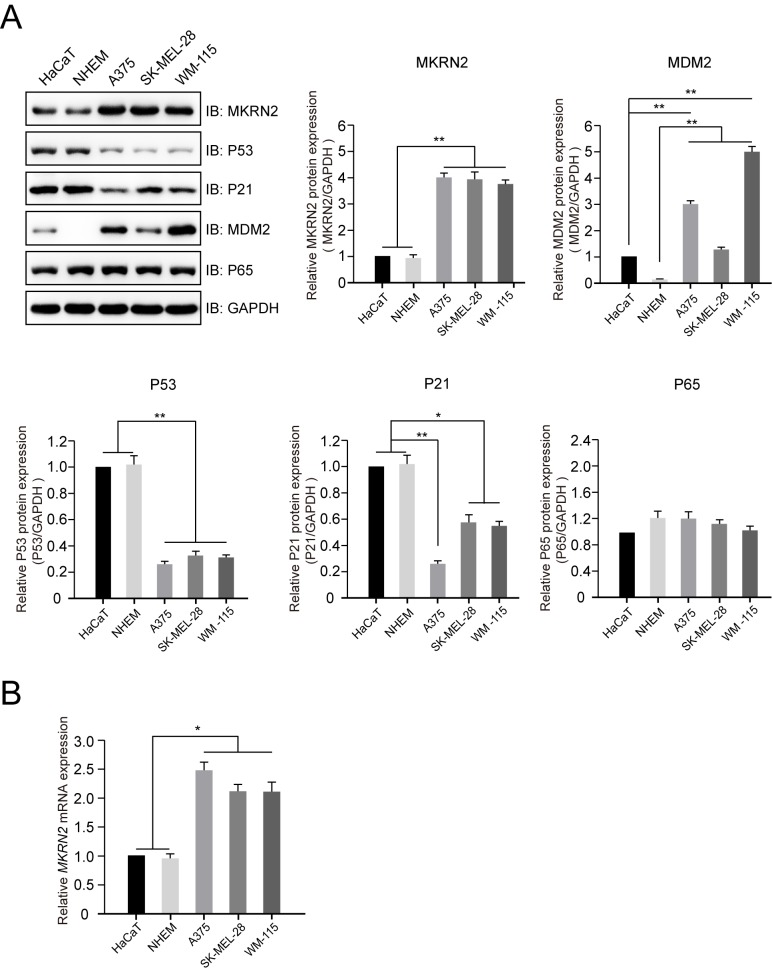
Figure 1.
Expression profile of MKRN2 in normal human epithelial melanocyte or keratinocyte and human malignant melanoma cell lines. (A) Protein expression profiles of MKRN2, P53, P21, MDM2 and P65 in normal human epithelial melanocyte (A375, SK-MEL-28 and WM-115), keratinocyte (HaCaT) and human malignant melanoma (NHEM) cell lines detected by immunoblotting. The protein expression levels of MKRN2, MDM2, P53, P21 and P65 in different cell lines were compared with those of GAPDH. n=3. (B) mRNA expression levels of MKRN2 in normal human epithelial melanocyte or keratinocyte cell lines and human malignant melanoma cell lines detected by reverse transcription-quantitative PCR. n=3. Data are expressed as the mean ± standard deviation and were analysed using one-way ANOVA with Tukey's post hoc test. *P<0.05, **P<0.01. MDM2, MDM2 proto-oncogene; MKRN2, makorin ring finger protein 2; P21, cyclin dependent kinase inhibitor 1A; P65, golgi reassembly stacking protein 1; IB, immunoblot.
Expression and purification of recombinant proteins
Firstly, the pGEX4T-1-Gst-MKRN2 (Ampicillin resistance) and pET28a-TP53-His6 (Kanamycin resistance) plasmids were expressed in BL21 E. coli. Monoclones were picked, cultured at 37°C in 2 ml lysogeny broth (LB) medium (10 g/l tryptone, 10 g/l yeast extract and 10 g/l NaCl) with the respective resistance overnight, then transferred to 1L LB medium and cultured for 5 h at 37°C under normal air conditions. Following isopropyl-β-d-thiogalactopyranoside (Sangon Biotech Co., Ltd.) induction at 16°C overnight the cells were lysed in PBS buffer and centrifuged at 4°C at 12,000 × g for 10 min, and incubated with glutathione or Ni2+ TA beads (GE Healthcare) to enrich the respective proteins. This was followed by elution with reduced 25 mM L-glutathione or 1 M imidazole dissolved in PBS buffer. The resulting products were dialyzed in PBS buffer supplemented with 20% glycerol prior to being aliquoted and preserved at −80°C.
Glutathione S-transferase (GST) pulldown assay
Purified Gst-MKRN2 (20 µg), P53-His6 (20 µg) and Glutathione Sepharose 4B were incubated at 4°C overnight in 500 µl pull-down buffer (20 mM Tris-Cl, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.5% (v/v) NP-40 and 10 µg/ml BSA (cat. no. SRE0096; Merck KGaA), pH 7.5). The beads were then centrifuged at 2,000 × g at 4°C for 2 min, and washed three times with pull-down buffer. Subsequently, the recovered beads were denatured at 100°C for 10 min in 2X SDS-PAGE loading buffer and subjected to immunoblotting analysis as aforementioned.
In vitro ubiquitination assay
In vitro ubiquitination assays were performed as previously described (21). Briefly, recombinant 200 ng His6-Ub, 200 ng UBA1-His6 (E1), 200 ng UBCH5A-His6 (E2), 500 ng Gst-MKRN2 (E3) and 500 ng P53-His6 were added to ubiquitination buffer (25 mM Tris-Cl, 100 mM NaCl, 1 mM DTT, 5 mM MgCl2; pH 7.6; supplemented with 2 mM ATP) with a final reaction volume of 50 µl and incubated at 37°C for 2 h. The ubiquitination levels of proteins were examined by an immunoblotting assay using an anti-P53 antibody as aforementioned.
P53-knockout cell line
The sgRNA targeting exon 1 of TP53 were designed and inserted into PX330 vector (cat. no. 98750; Addgene, Inc.). A P53−/− knockout cell line was established using the CRISPR-Cas9 technique, as previously described (18). Briefly, A375 cells (100,000 cells/well) were transfected with 3 µg CRISPR-Cas9-based sgRNA (PX330-P53-sgRNA), and monoclonal cells were chosen and detected by immunoblotting analysis. Subsequently, genetic ablation of TP53 was confirmed by first generation sequencing.
Statistical analysis
All experiments were performed in triplicate. All values are presented as the mean ± SD. One-way ANOVA followed by Tukey's post hoc multiple comparisons test was performed using GraphPad Prism v7 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference, whereas P<0.01 was considered to indicate a statistically very significant difference.
Results
Higher expression of MKRN2 in human malignant melanoma cell lines
To investigate the expression profile of MKRN2 in human malignant melanoma cell lines, lysates of three melanoma cell lines (A375, SK-MEL-28 and WM-115) and two normal cell lines (HaCaT, immortal human keratinocyte cell line; and NHEM, primary-cultured normal human epithelial melanocyte cell line) were prepared. Immunoblotting analysis indicated that the protein levels of MKRN2 were significantly higher in the three melanoma cell lines compared with in the two normal cell lines (Fig. 1A), which was consistent with the mRNA levels of MKRN2 detected by RT-qPCR (Fig. 1B). The protein expression levels of P53 and P21 were higher in the two normal cell lines compared with in the three melanoma cell lines. P65, a substrate for E3 ligase for MKRN2, exhibited weak alterations in these five cell lines. Additionally, MDM2, an E3 ligase for P53, expression was higher in the three melanoma cell lines compared with in the two normal cell lines (Fig. 1A). These results suggested that MKRN2 exhibited higher expression in human malignant melanoma cells and its expression was negatively associated with P53 and P21 expression.
Downregulation of MKRN2 inhibits melanoma cell growth
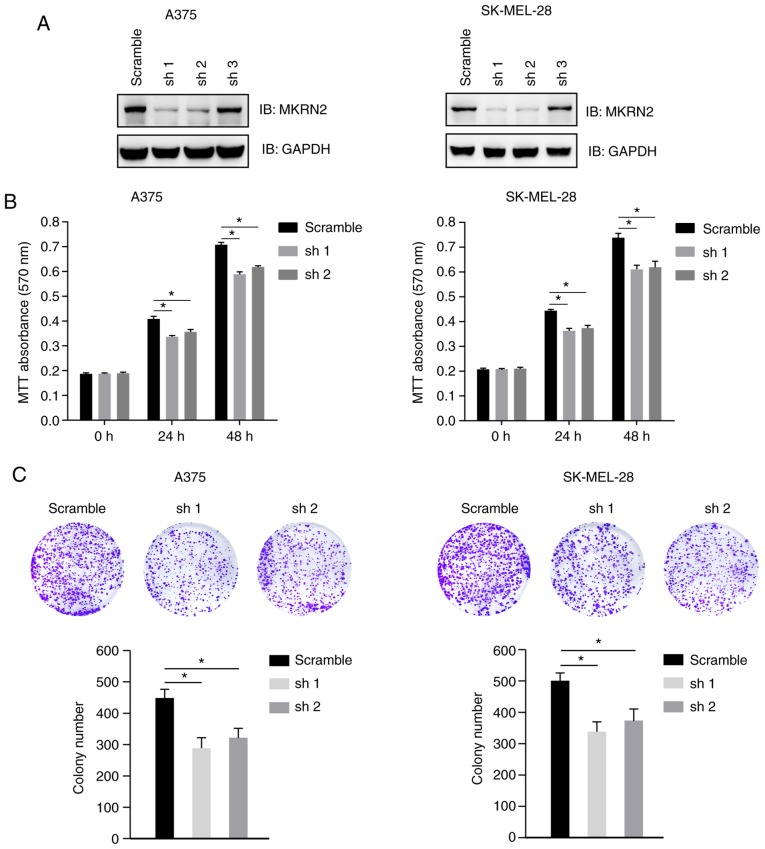
To investigate the effect of MKRN2 on melanoma cell proliferation, three MKRN2 shRNAs were designed and tested in two melanoma cell lines (A375 and SK-MEL-28). These two cell lines were selected for further study, as the expression of MKRN2 was higher in these two cell lines compared with that in WM-115. Immunoblotting analysis indicated that MKRN2 expression was markedly decreased in cells stably transfected with shRNA1 and shRNA2, and these cells were selected for further study (Fig. 2A). Cell viability of A375 and SK-MEL-28 cells was detected by MTT assay at 24 and 48 h post culture, and significant cell growth arrest was observed in MKRN2-knockdown cell lines compared with in the control group (Fig. 2B). Decreased colony numbers were identified in MKRN2-knockdown groups compared with in the control group in A375 and SK-MEL-28 cells (Fig. 2C). These results suggested that downregulation of MKRN2 inhibited melanoma cell growth.
Figure 2.
MKRN2-knockdown induces arrest of melanoma cell growth. (A) Test of the knockdown efficiency of shRNAs targeting MKRN2. A375 and SK-MEL-28 cells were transfected with MKRN2 shRNAs and then subjected to screening for puromycin resistance to establish stably expressed cell lines. (B) MKRN2-knockdown inhibited melanoma cell proliferation. A375 and SK-MEL-28 cells that stably expressed MKRN2 shRNAs were seeded into a 96-well plate and detected using an MTT assay at the indicated time points. The 0 h time point was defined as 6 h after the cells were seeded. n=3. (C) MKRN2-knockdown inhibited melanoma cell colony formation. A375 and SK-MEL-28 cells that stably expressed MKRN2 shRNAs were seeded into a 6-well plate; colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet 7 days later. The images were obtained using a camera, colony numbers were counted and the average number of colonies was calculated. Three samples were analysed for each group. Data are expressed as the mean ± standard deviation and were analysed using one-way ANOVA with Tukey's post hoc test. *P<0.05. MKRN2, makorin ring finger protein 2; sh, short hairpin RNA; shRNA, short hairpin RNA; IB, immunoblot.
Downregulation of MKRN2 inhibits melanoma cell growth by upregulating P53
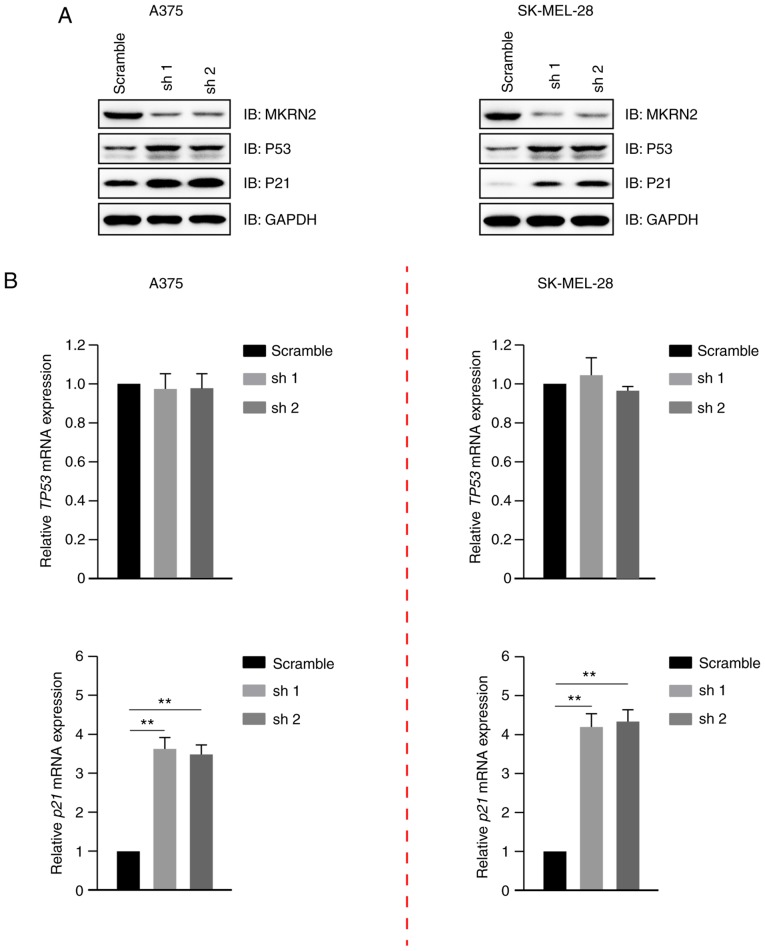
MKRN2 expression exhibited a negative association with P53 and P21 expression (Fig. 1A), and its effect on these was detected. Immunoblotting analysis indicated that P53 and P21 expression were markedly increased in MKRN2-knockdown cells compared with in the control A375 and SK-MEL-28 cells (Fig. 3A). It has been widely reported that P53 is a transcriptional activator of the p21 gene (22,23). The gene expression levels of TP53 and p21 were detected by RT-qPCR, which revealed that the mRNA levels of p21, but not TP53, were upregulated in MKRN2-knockdown cells (Fig. 3B). These data indicated that MKRN2-knockdown increased the stability of the P53 protein and had little effect on its mRNA levels.
Figure 3.
MKRN2-knockdown induces the arrest of melanoma cell growth by upregulating P53. (A) Protein expression levels of P53 and P21 were upregulated in MKRN2-knockdown cells. The protein levels of P53 and P21 were detected by immunoblotting in melanoma cell lines (A375 and SK-MEL-28) that stably expressed MKRN2 shRNAs. (B) mRNA levels of P21 but not P53 were upregulated in MKRN2-knockdown cells. The mRNA levels of TP53 and p21 were detected by reverse transcription-quantitative PCR in melanoma cells (A375 and SK-MEL-28) that stably expressed MKRN2 shRNAs. Data are expressed as the mean ± standard deviation and were analysed using one-way ANOVA with Tukey's post hoc test. n=3. **P<0.01. MKRN2, makorin ring finger protein 2; P21, cyclin dependent kinase inhibitor 1A; sh, short hairpin; sh, short hairpin RNA; shRNA, short hairpin RNA; IB, immunoblot.
MKRN2 interacts with and ubiquitylates P53
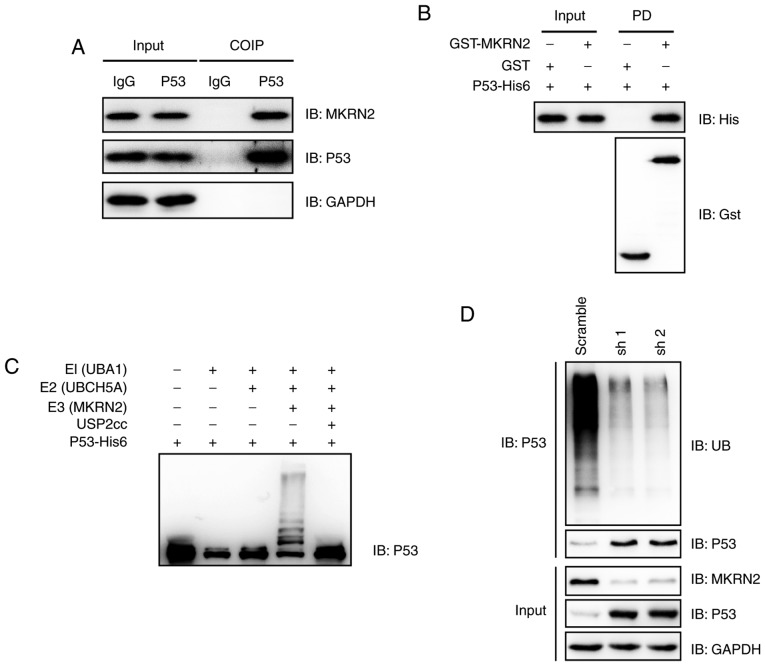
The observations that MKRN2 affected the growth rate of melanoma cells by affecting P53 protein levels prompted the present study to determine whether P53 and MKRN2 could interact with each other. MKRN2 expression was higher in the A375 cell line compared with that in SK-MEL-28 and WM-115 cells; therefore A375 was selected for further study. Immunoprecipitation analyses demonstrated that endogenous MKRN2 formed a complex with endogenous P53 in A375 cells (Fig. 4A). Subsequently, the present study assessed whether MKRN2 could directly interact with P53 in vitro. Recombinant GST-tagged MKRN2 and His6-tagged P53 were purified, and then GST pull-down assays were performed. These assays demonstrated that MKRN2 and P53 formed a complex in vitro (Fig. 4B). An in vitro ubiquitylation assay was carried out with recombinant proteins, and immunoblotting analysis indicated that MKRN2 ubiquitylated P53 (Fig. 4C). Additionally, the present study indicated that ablation of MKRN2 reduced the ubiquitylation levels of P53 in A375 cells (Fig. 4D). Overall, these data suggested that MKRN2 interacted with and ubiquitylated P53.
Figure 4.
MKRN2 interacts with and ubiquitylates P53. (A) Endogenous MKRN2 and P53 formed a complex in A375 cells. A375 cell lysates were immunoprecipitated with anti-P53 antibody and subjected to immunoblotting analysis. (B) MKRN2 interacted with P53 in vitro. Recombinant GST-tagged MKRN2 and His6-tagged P53 were purified, and then GST pull-down assays were performed and subjected to immunoblotting analysis. (C) MKRN2 ubiquitylated P53 in vitro. An in vitro ubiquitylation assay was carried out with the recombinant proteins P53-His6, E1 (UBA1-His6), E2 (UBCH5A-His6), His6-Ub and Gst-MKRN2, together with the indicated components, and subjected to immunoblotting analysis. (D) MKRN2 ubiquitylated P53 in vivo. A375 cells stably transfected with MKRN2 shRNAs were lysed in RIPA buffer and immunoprecipitated with anti-P53 antibody and subjected to immunoblotting analysis using the indicated antibodies. COIP, co-immunoprecipitation; GST, glutathione S-transferase; His, histidine; IgG, immunoglobulin G; MKRN2, makorin ring finger protein 2; sh, short hairpin RNA; shRNA, short hairpin RNA; UBA1, ubiquitin like modifier activating enzyme 1; UBCH5A, ubiquitin conjugating enzyme E2 D1; USP2, ubiquitin specific peptidase 2; IB, immunoblot.
MKRN2 regulates the proliferation of melanoma cells in a P53-dependent manner
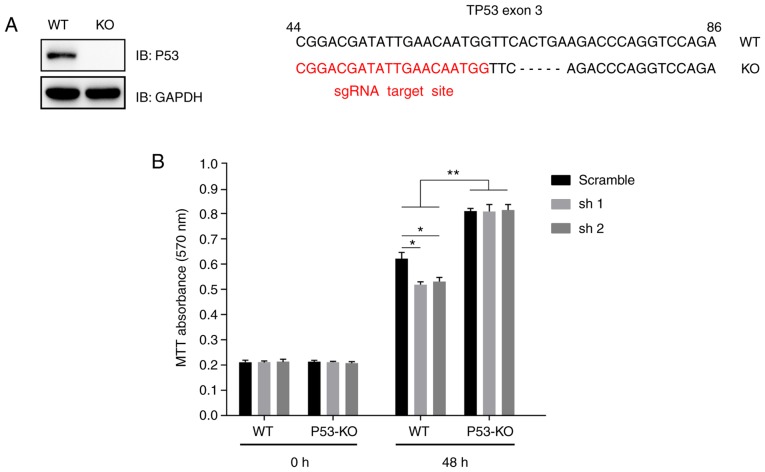
As MKRN2 expression was higher in the A375 cell line compared with that in SK-MEL-28 and WM-115 cells, these were selected for further study. A TP53-knockout (TP53−/−) A375 cell line was generated using the CRISPR-Cas9 sgRNA-based method and was validated by immunoblotting analysis (Fig. 5A). First-generation sequencing indicated that the knockout cell line had a 5-bp deletion on exon 3 of the TP53 genomic DNA (Fig. 5A). Wild type or TP53−/− A375 cells that stably expressed MKRN2 shRNAs were seeded into a 96-well plate, and cell viability was detected using an MTT assay at 48 h. The results indicated that MKRN2-knockdown induced growth arrest in wild type A375 cells but had little effect in TP53−/− A375 cells. Additionally, P53-knockout significantly promoted A375 cell proliferation, independent of whether MKRN2 was knocked down (Fig. 5B).
Figure 5.
MKRN2 regulates the proliferation of melanoma cells in a P53-dependent manner. (A) A TP53−/− A375 cell line was established using the CRISPR-Cas9 technique. A375 cells were transfected with CRISPR-Cas9-based sgRNA indicated in red (right panel), monoclonal cells were picked and detected by immunoblotting analysis (left panel), and then genetic ablation of TP53 with a 5-bp deletion was confirmed by sequencing. (B) MKRN2 knockdown induced melanoma cell growth arrest in a P53-dependent manner. WT or TP53−/− A375 cells that stably expressed MKRN2 shRNAs were seeded into a 96-well plate and detected by MTT assay at the indicated time points (0 and 48 h). The 0 h time point was defined as 6 h after the cells were seeded. Data are expressed as the mean ± standard deviation and were analysed using one-way ANOVA with Tukey's post hoc test. n=3. *P<0.05, **P<0.01. KO, knockout; MKRN2, makorin ring finger protein 2; sgRNA, single guide RNA; sh, short hairpin RNA; shRNA, short hairpin RNA; WT, wild-type; IB, immunoblot.
Discussion
The present study demonstrated that the protein expression levels of MKRN2 were higher in melanoma cells and were negatively associated with P53 and P21 expression. Furthermore, downregulation of MKRN2 induced the arrest of melanoma cell growth, which suggested that MKRN2 may act as an oncogene. In addition, the present study identified that MKRN2 acted as a novel E3 ligase for P53 and reduced the stability of the P53 protein. Finally, a P53-knockout cell line was established, and it was demonstrated that MKRN2 regulated the proliferation of melanoma cells in a P53-dependent manner.
MKRN2 belongs to the MAKORIN family, which includes three proteins: Makorin ring finger protein 1 (MKRN1), MKRN2 and makorin ring finger protein 3 (24). MAKORIN proteins share a highly homologous amino acid sequence, particularly in the zinc finger domains, suggesting that they may share similar functions or regulatory mechanisms (24). The function of MKRN1 has been explored previously, and MKRN1 acts as an E3 ubiquitin ligase, inducing degradation of human telomerase reverse transcriptase, viral capsid proteins, p53 and p21 cell cycle regulators, peroxisome proliferator activated receptor γ, adenomatous polyposis coli and AMP-activated protein kinase α1 (24–27). Similar to MKRN1, the present study identified that MKRN2 interacted with and ubiquitylated P53 (27); however, MKRN2 had no direct effect on P21 protein stability.
A previous study has demonstrated that MKRN2 inhibits the migration and invasion of non-small-cell lung cancer by negatively regulating the PI3K/Akt signalling pathway (12), suggesting that MKRN2 acts as a tumour suppressor in lung cancer. This finding contradicts the present study, which indicated that MKRN2 acts as an oncogene in melanoma. However, a study by Lee et al (28) demonstrated that overexpression of MKRN2 increases the proliferation of K562 cells, which is consistent with the present study. Whether MKRN2 acts as a tumour suppressor gene or oncogene may depend on the cancer type.
MKRN2 has been identified as an E3 ligase for the p65 subunit of NF-κB and negatively regulates inflammatory responses (17). However, P65 exhibited weak alterations in the three melanoma cell lines compared with in the two normal cell lines used in the present study. This result may suggest that the functions of MKRN2 depend on cell/cancer types.
P53 is the most well-known tumour suppressor, and >20 E3s have been reported, including MDM2, ubiquitin protein ligase E3A and MKRN1 (27,29). The ubiquitination and degradation of P53 serves an important role in cell cycle regulation and tumorigenesis; the protein expression levels of MKRN2 and MDM2 were higher in melanoma cells. Additionally, MDM2 is a well-known proto-oncogene in numerous types of cancer, such as osteosarcoma, glioblastoma and breast cancer, and it is an important target for cancer therapy (30,31). Therefore, one may hypothesize that MKRN2 may also be an important marker for melanoma and act as a potential target for its therapy. Testing of MKRN2 function in animal models and patient samples will be conducted in future studies.
Acknowledgements
The authors would like to thank Professor Ronggui Hu (State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) for his technical support and providing the proteins E1 [ubiquitin like modifier activating enzyme 1 (UBA1)-hexahistidine (His6)], E2 [ubiquitin conjugating enzyme E2 D1 (UBCH5A)-His6] and His6-Ub, the plasmids pET28a-TP53-His6 and pGEX4T-1-Gst-MKRN2, and the cell lines (HaCaT and NHEM).
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during the present study are included in this published article.
Authors' contributions
GZ and YZ conceived and designed the experiments. YZ and NC performed the experiments, collected the data and analysed the results. GZ and YZ wrote the paper.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chu H, Li M, Wang X. Capsaicin induces apoptosis and autophagy in human melanoma cells. Oncol Lett. 2019;17:4827–4834. doi: 10.3892/ol.2019.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra H, Mishra PK, Ekielski A, Jaggi M, Iqbal Z, Talegaonkar S. Melanoma treatment: From conventional to nanotechnology. J Cancer Res Clin Oncol. 2018;144:2283–2302. doi: 10.1007/s00432-018-2726-1. [DOI] [PubMed] [Google Scholar]
- 3.Martens MC, Seebode C, Lehmann J, Emmert S. Photocarcinogenesis and skin cancer prevention strategies: An update. Anticancer Res. 2018;38:1153–1158. doi: 10.21873/anticanres.12334. [DOI] [PubMed] [Google Scholar]
- 4.Rick JW, Shahin M, Chandra A, Dalle Ore C, Yue JK, Nguyen A, Yagnik G, Sagar S, Arfaie S, Aghi MK. Systemic therapy for brain metastases. Crit Rev Oncol Hematol. 2019;142:44–50. doi: 10.1016/j.critrevonc.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhary M, Patel KR, Danish HH, Lawson DH, Khan MK. BRAF inhibitors and radiotherapy for melanoma brain metastases: Potential advantages and disadvantages of combination therapy. Onco Targets Ther. 2016;9:7149–7159. doi: 10.2147/OTT.S119428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholas S, Mathios D, Jackson C, Lim M. Metastatic melanoma to the brain: Surgery and radiation is still the standard of care. Curr Treat Options Oncol. 2013;14:264–279. doi: 10.1007/s11864-013-0228-6. [DOI] [PubMed] [Google Scholar]
- 7.Mandala M, Voit C. Targeting BRAF in melanoma: Biological and clinical challenges. Crit Rev Oncol Hematol. 2013;87:239–255. doi: 10.1016/j.critrevonc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrescu DT, Ichim TE, Riordan NH, Marincola FM, Di Nardo A, Kabigting FD, Dasanu CA. Immunotherapy for melanoma: Current status and perspectives. J Immunother. 2010;33:570–590. doi: 10.1097/CJI.0b013e3181e032e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimanovsky A, Jethava A, Dasanu CA. Immune alterations in malignant melanoma and current immunotherapy concepts. Expert Opin Biol Ther. 2013;13:1413–1427. doi: 10.1517/14712598.2013.827658. [DOI] [PubMed] [Google Scholar]
- 10.Walter L, Heinzerling L. BRAF Inhibitors and radiation do not act synergistically to inhibit WT and V600E BRAF human melanoma. Anticancer Res. 2018;38:1335–1341. doi: 10.21873/anticanres.12356. [DOI] [PubMed] [Google Scholar]
- 11.Gray TA, Azama K, Whitmore K, Min A, Abe S, Nicholls RD. Phylogenetic conservation of the makorin-2 gene, encoding a multiple zinc-finger protein, antisense to the RAF1 proto-oncogene. Genomics. 2001;77:119–126. doi: 10.1006/geno.2001.6627. [DOI] [PubMed] [Google Scholar]
- 12.Jiang J, Xu Y, Ren H, Wudu M, Wang Q, Song X, Su H, Jiang X, Jiang L, Qiu X. MKRN2 inhibits migration and invasion of non-small-cell lung cancer by negatively regulating the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37:189. doi: 10.1186/s13046-018-0855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 15.Gray TA, Hernandez L, Carey AH, Schaldach MA, Smithwick MJ, Rus K, Marshall Graves JA, Stewart CL, Nicholls RD. The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics. 2000;66:76–86. doi: 10.1006/geno.2000.6199. [DOI] [PubMed] [Google Scholar]
- 16.Borden KL. RING domains: Master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 17.Shin C, Ito Y, Ichikawa S, Tokunaga M, Sakata-Sogawa K, Tanaka T. MKRN2 is a novel ubiquitin E3 ligase for the p65 subunit of NF-κB and negatively regulates inflammatory responses. Sci Rep. 2017;7:46097. doi: 10.1038/srep46097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Li C, Gao X, Xia K, Guo H, Li Y, Hao Z, Zhang L, Gao D, Xu C, et al. Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res. 2018;28:48–68. doi: 10.1038/cr.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Chen P, Zhang J, Zhang L, Huang X, Yao Y, Che X, Fan X, Ge S, Wang Z. Enzyme-induced vitreolysis can alleviate the progression of diabetic retinopathy through the HIF-1α pathway. Invest Ophthalmol Vis Sci. 2013;54:4964–4970. doi: 10.1167/iovs.12-11443. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 23.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 24.Abreu AP, Macedo DB, Brito VN, Kaiser UB, Latronico AC. A new pathway in the control of the initiation of puberty: The MKRN3 gene. J Mol Endocrinol. 2015;54:R131–R139. doi: 10.1530/JME-14-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HK, Lee EW, Seo J, Jeong M, Lee SH, Kim SY, Jho EH, Choi CH, Chung JY, Song J. Ubiquitylation and degradation of adenomatous polyposis coli by MKRN1 enhances Wnt/β-catenin signaling. Oncogene. 2018;37:4273–4286. doi: 10.1038/s41388-018-0267-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee MS, Han HJ, Han SY, Kim IY, Chae S, Lee CS, Kim SE, Yoon SG, Park JW, Kim JH, et al. Loss of the E3 ubiquitin ligase MKRN1 represses diet-induced metabolic syndrome through AMPK activation. Nat Commun. 2018;9:3404. doi: 10.1038/s41467-018-05721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, Song J. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KY, Chan KY, Tsang KS, Chen YC, Kung HF, Ng PC, Li CK, Leung KT, Li K. Ubiquitous expression of MAKORIN-2 in normal and malignant hematopoietic cells and its growth promoting activity. PLoS One. 2014;9:e92706. doi: 10.1371/journal.pone.0092706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sane S, Rezvani K. Essential roles of E3 ubiquitin ligases in p53 regulation. Int J Mol Sci. 2017;18(pii):E442. doi: 10.3390/ijms18020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta A, Shah K, Oza MJ, Behl T. Reactivation of p53 gene by MDM2 inhibitors: A novel therapy for cancer treatment. Biomed Pharmacother. 2019;109:484–492. doi: 10.1016/j.biopha.2018.10.155. [DOI] [PubMed] [Google Scholar]
- 31.Niazi S, Purohit M, Niazi JH. Role of p53 circuitry in tumorigenesis: A brief review. Eur J Med Chem. 2018;158:7–24. doi: 10.1016/j.ejmech.2018.08.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during the present study are included in this published article.