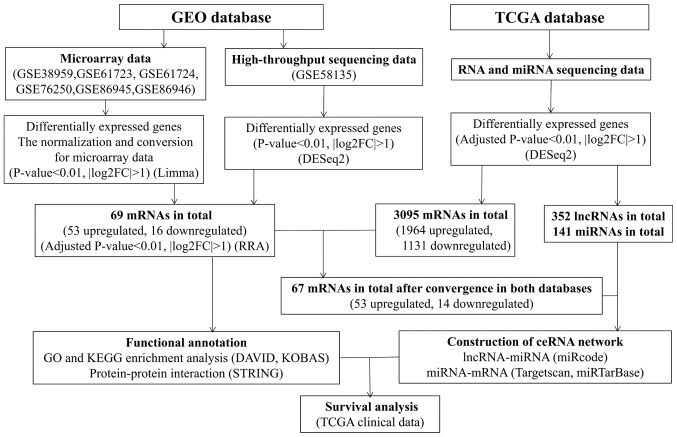
Figure 1.
Flowchart of data collection and method implementation in this study. GEO, Gene Expression Omnibus; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; TCGA, The Cancer Genome Atlas; DAVID, Database for Annotation, Visualization and Integrated Discovery; STRING, Search Tool for the Retrieval of Interacting Genes; KOBAS, KEGG Orthology Based Annotation System; FC, fold change; miRNAs, microRNAs; lncRNAs, long non-coding RNAs; ceRNA, competing endogenous RNA.