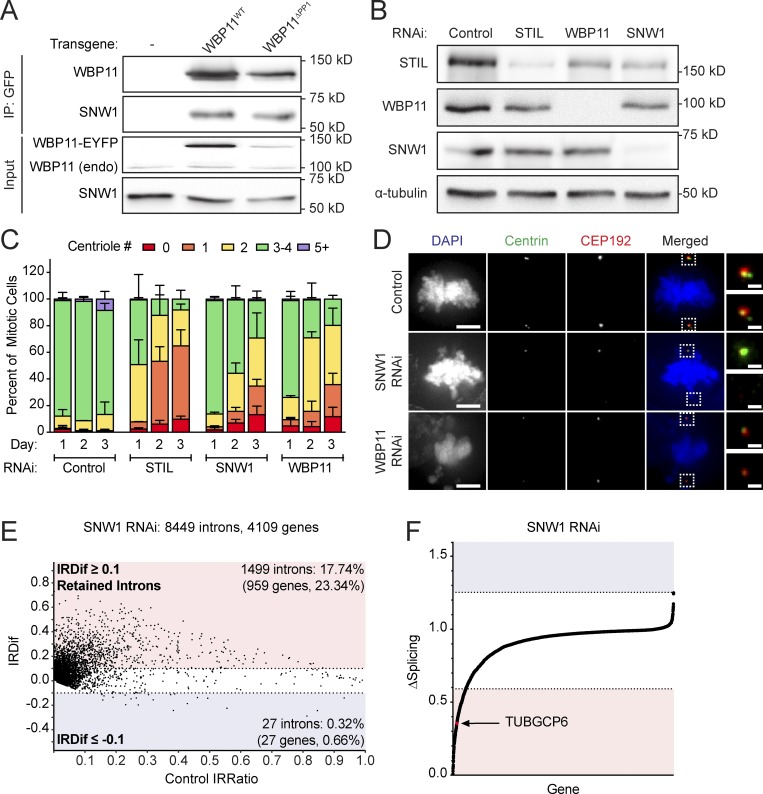
Figure 5.
SNW1 interacts with WBP11 and is required for centriole biogenesis. (A) Immunoblot showing coimmunoprecipitation (IP) of endogenous SNW1 with WBP11WT-EYFP and WBP11ΔPP1-EYFP. (B) Immunoblot showing expression levels of STIL, SNW1, and WBP11 72 h after siRNA transfection. (C) Quantification of centriole number in mitotic cells 72 h after siRNA-mediated depletion of SNW1. Data are shown alongside the data for WBP11 and STIL from Fig. 1 B. n = 3, ≥49 cells per experiment. Error bars represent SD. (D) Representative images of cells from C transfected with a WBP11 or SNW1 siRNA. Scale bars represent 5 µm; 1 µm in zoomed-in regions. (E) Classification of introns detected by mRNA-seq 72 h after SNW1 depletion by RNAi. The IRDif score of each intron was plotted against the IRratio of that same intron in the control sample. Colors indicate introns that are excised more efficiently (blue), or retained more often (red), following SNW1 depletion. Raw data are shown in Table S5. (F) Plot showing the change in the fraction of completely spliced mRNA for each gene 72 h after SNW1 depletion with siRNA. Colors indicate three SDs above (blue) or below (red) the average level of mRNA splicing observed. Raw data are shown in Table S6.