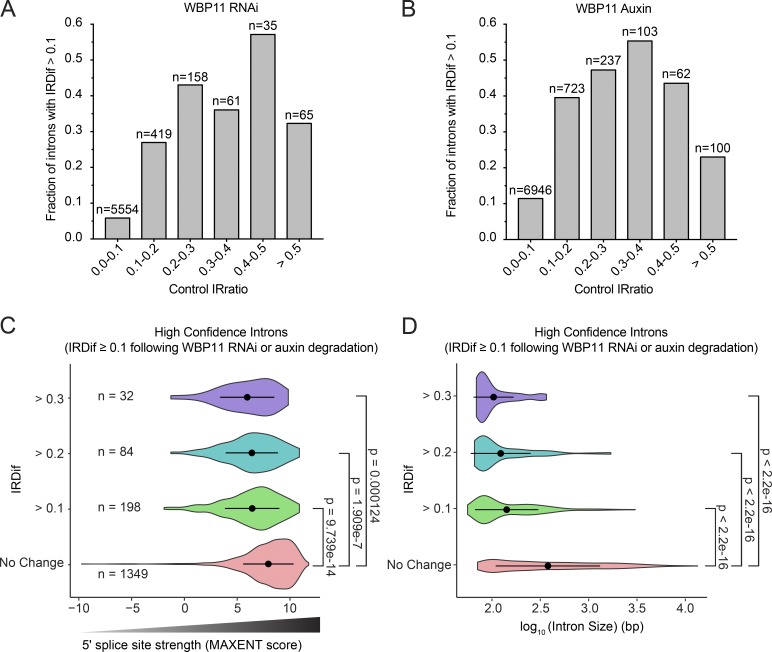
Figure 9.
WBP11 is required for splicing short introns with weak 5′ splice sites. (A and B) Introns from the WBP11 RNAi RNA-seq experiment (A) or the WBP11 auxin RNA-seq experiment (B) were divided into six bins based on their IRratio in the control sample. For each bin, we calculated the fraction of introns that required WBP11 for efficient splicing (IRDif score ≥0.1). The probability an intron would be retained generally increased as the control IRratio increased from 0 to 0.5. Therefore, introns that are weakly spliced in the control sample are more likely to be retained following WBP11 depletion. (C) Violin plots of MAXENT splice site scores of introns that are significantly retained following WBP11 RNAi and degradation with auxin. Introns were divided into groups based on their IRDif scores. No change = IRDif ≥−0.1 and ≤0.1. Introns that are more highly retained (larger IRDif values) following depletion of WBP11 have weaker splice site scores (lower MAXENT values). Points represent the mean and bars display the SD. P values were calculated using an unpaired parametric t test. (D) Violin plots showing the length of the introns that are significantly retained following WBP11 RNAi and degradation with auxin. Introns were divided into groups based on their IRDif scores. No change = IRDif ≥−0.1 and ≤0.1. Introns that are more highly retained (larger IRDif values) following depletion of WBP11 are smaller in size. Points represent the mean and bars display the SD. P values were calculated using an unpaired parametric t test.