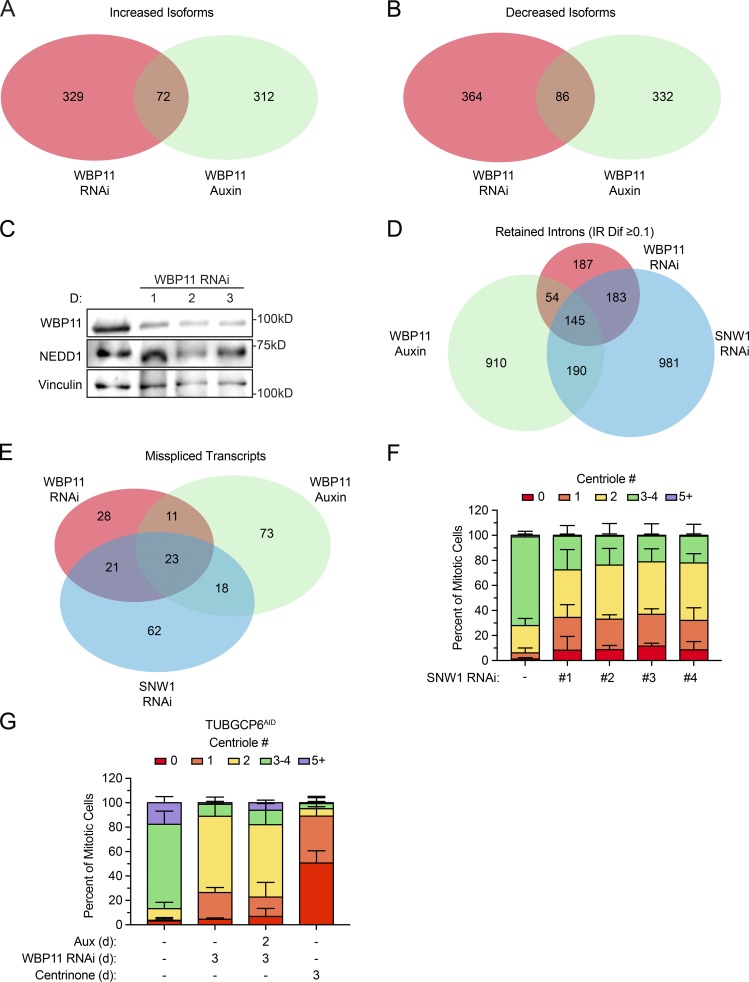
Figure S4.
WBP11 and SNW1 are required for the correct splicing of a common set of pre-mRNAs. (A) Overlapping hits for isoforms with expression increased more than two SDs following WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 72 isoforms that depend on both WBP11 and SNW1 for appropriate expression were identified. (B) Overlapping hits for isoforms with expression decreased more than two SDs following WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 86 isoforms that depend on both WBP11 and SNW1 for appropriate expression were identified. (C) Immunoblot showing NEDD1 protein levels after siRNA-mediated depletion of WBP11. (D) Overlapping hits for retained introns with an IRDif score of ≥0.1 following WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 145 introns that depend on WBP11 and SNW1 for efficient splicing were identified. (E) Overlapping hits for the genes that experienced a greater than three SD decrease of correctly spliced mRNA after WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 23 genes that depend on WBP11 and SNW1 for efficient splicing were identified. (F) Quantification of centriole number in mitotic cells 72 h after depletion of SNW1 with one of four independent siRNAs. n = 3, ≥50 cells per experiment. Error bars represent SD. (G) Quantification of centriole number in mitotic cells 72 h after depletion of WBP11 by siRNA in TUBGCP6AID cells. TUBGCP6AID was degraded by auxin where indicated. n = 3, ≥50 cells per experiment. Error bars represent SD.