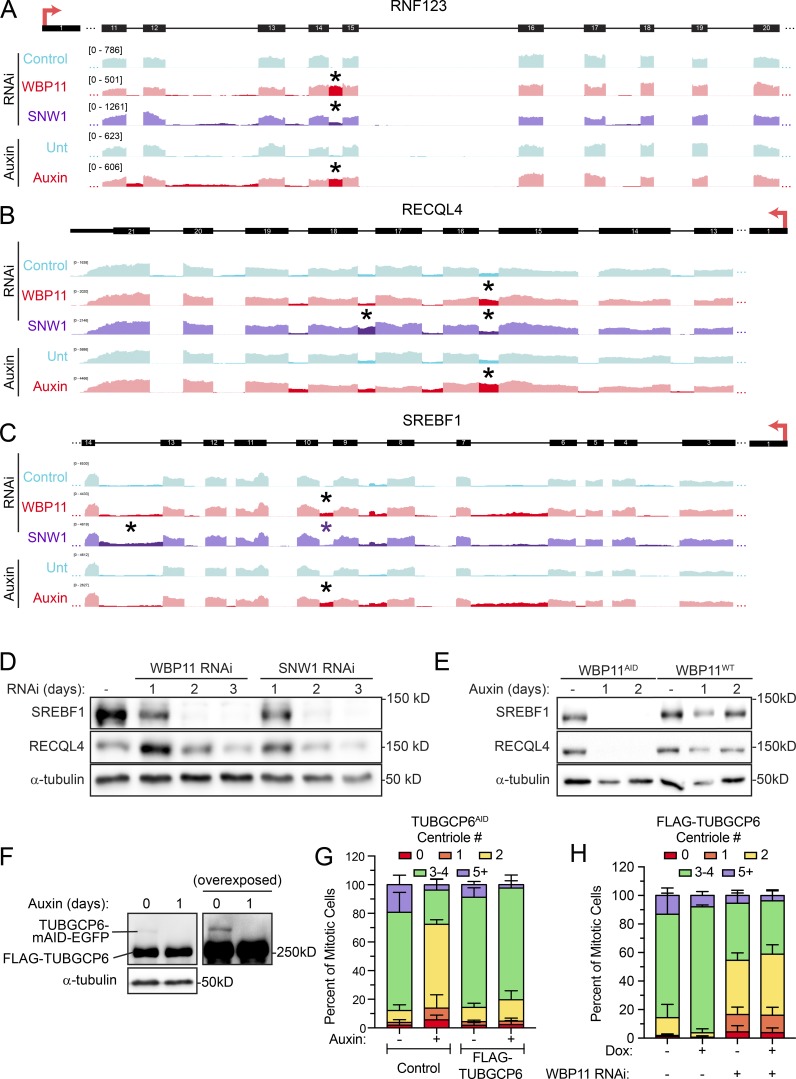
Figure S5.
WBP11 and SNW1 are required for the correct splicing of a subset of introns. (A) A region of the RNF123 gene is aligned to IGV reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. RNF123 requires WBP11 and, to a lesser extent, SNW1 for efficient splicing of one of its introns (*). (B) A region of the RECQL4 gene is aligned to IGV reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. RECQL4 requires WBP11 and SNW1 for efficient splicing of some introns (*). Note that some introns require SNW1, but not WBP11, for efficient splicing. (C) A region of the SREBF1 gene is aligned to IGV reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. SREBF1 requires WBP11 and SNW1 for efficient splicing of some introns (*). Note that some introns require SNW1, but not WBP11, for efficient splicing. (D) Immunoblot showing expression levels of RECQL4 and SREBF1 after 72 h of SNW1 or WBP11 siRNA knockdown. (E) Immunoblot showing expression levels of RECQL4 and SREBF1 in WBP11WT and WBP11AID cells at 48 h after auxin addition. (F) Immunoblot showing TUBGCP6 expression in TUBGCP6AID cells expressing a FLAG-TUBGCP6 transgene. (G) Quantification of centriole number in mitotic TUBGCP6AID cells 24 h after auxin addition. TUBGCP6AID cells with or without expression of a FLAG-TUBGCP6 transgene were used. Data are shown alongside the TUBGCP6AID data from Fig. 7 C for comparison. n = 3, 50 cells per experiment. Error bars represent SD. (H) Graph showing the percentage of mitotic cells with three to four centrioles after 72 h of WBP11 depletion by siRNA. Expression of the FLAG-TUBGCP6 was induced by addition of doxycycline where indicated. n = 3, 50 cells/experiment. Error bars represent SD.