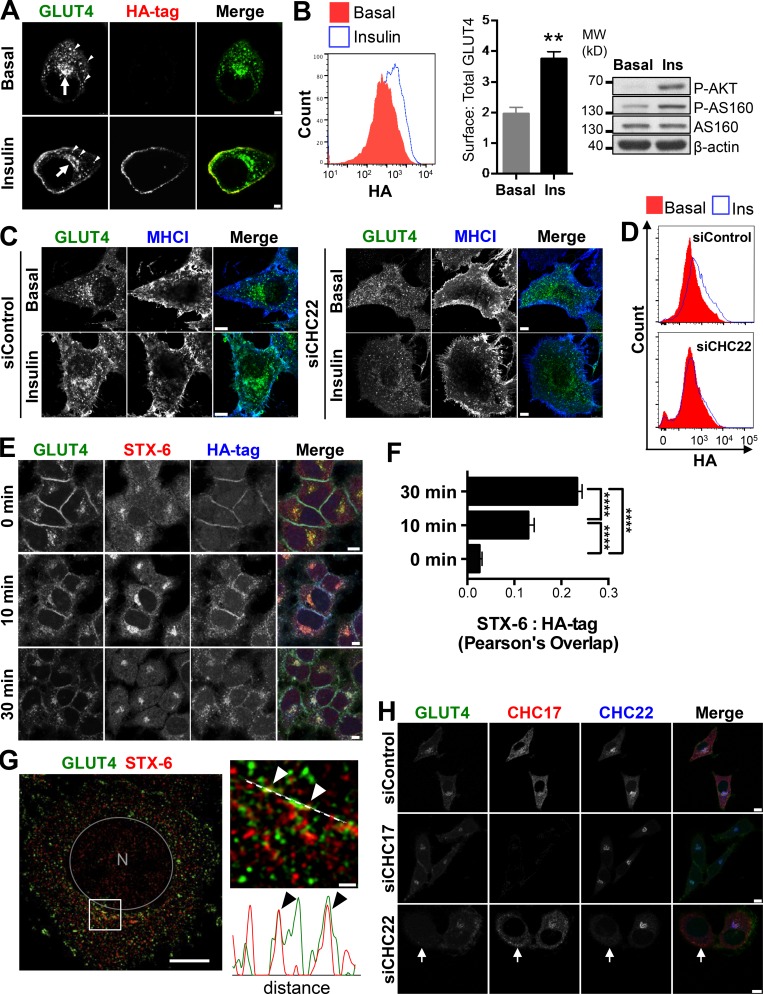
Figure 1.
HeLa-GLUT4 cells have a functional GLUT4 trafficking pathway that requires CHC22. (A) Representative images of GLUT4 (exofacial HA tag, internal GFP tag) in HeLa-GLUT4 cells before (basal) or after insulin treatment. GLUT4 at the plasma membrane was detected by IF after surface labeling with anti-HA monoclonal antibody (red). Total GLUT4 (green) was detected by GFP tag. Arrows show the GSC. Arrowheads point to peripheral GLUT4 vesicles. Scale bars: 7.5 µm. (B) Left: Representative FACS histogram of surface GLUT4 fluorescence intensities (signal from anti-HA labeling) before (basal) and after insulin treatment (Ins). Middle: Quantification of surface-to-total GLUT4 (HA-to-GFP MFI signals). Data expressed as mean ± SEM, n = 3, 10,000 cells acquired per experiment. Two-tailed unpaired Student’s t test with equal variances: **, P < 0.01. Right: Representative immunoblot for phosphorylated AKT (p-AKT), phosphorylated AS160 (p-AS160), total AS160, and β-actin in HeLa-GLUT4 cells before and after insulin treatment. The migration position of molecular weight (MW) markers is indicated at the left in kilodaltons. (C) Representative images of total GLUT4 (GFP tag, green) and MHCI (blue) before (basal) or after insulin treatment in HeLa-GLUT4 cells transfected with nontargeting control siRNA (siControl) or siRNA targeting CHC22 (siCHC22). Scale bars: 8 µm. (D) Representative FACS histograms of surface GLUT4 fluorescence intensity (signal from anti-HA labeling) in HeLa-GLUT4 cells transfected with siControl or siRNA siCHC22 before (red) or after (blue) treatment with insulin. Histograms are extracted from the experiment quantified in Fig. 8 A. (E) Representative IF staining for internalized surface-labeled GLUT4 (HA tag, blue) and STX-6 (red) for HeLa-GLUT4 cells at 0, 10, or 30 min after insulin treatment. Total GLUT4 is detected by GFP tag (green). Scale bars: 7.5 µm. (F) Pearson’s overlap quantification for labeling of STX-6 and HA tag. Data expressed as mean ± SEM, n = 3, 14–19 cells per experiment. One-way ANOVA followed by Bonferroni’s multiple comparison post hoc test, ****, P < 0.0001. (G) Left: Representative SIM image of a HeLa-GLUT4 cell stained for STX-6 (red). Total GLUT4 (green) was detected by GFP tag. The gray circle delineates the nucleus (N) and the white square delineates the magnified area displayed in the right image. Scale bars: 10 µm; magnified image: 1 µm. Right: The white dashed line in the magnified area spans the segment for which fluorescence intensities for GLUT4 and STX-6 are plotted below, in green and red, respectively. Arrowheads indicate areas of overlap. (H) Representative IF staining for CHC17 (red) and CHC22 (blue) in HeLa-GLUT4 cells transfected with nontargeting siControl or siRNA targeting CHC17 (siCHC17) or siCHC22, with GLUT4 detected by GFP tag (green). Arrows point to a CHC22-depleted cell. Scale bars: 10 µm for siControl and siCHC17 and 7.5 µm for siCHC22. Merged images in A, C, E, G, and H show red/green overlap in yellow, red/blue overlap in magenta, green/blue overlap in cyan, and red/green/blue overlap in white.