Figure 4.
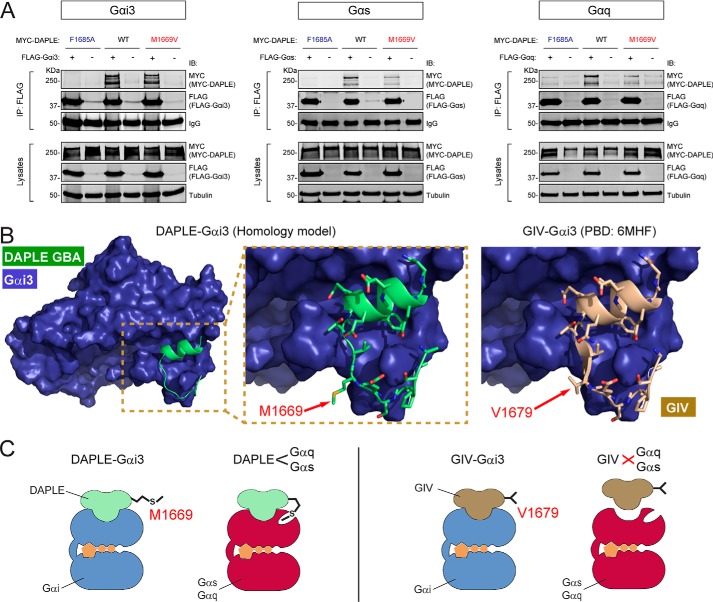
M1669V mutation in full-length DAPLE disrupts binding to Gαs or Gαq but not to Gαi3. A, co-immunoprecipitation experiments comparing the effect of DAPLE M1669V and F1675A mutations on G-protein binding, which show that the former disrupts binding to Gαs and Gαq, but not to Gαi3, whereas the latter disrupts binding to all G proteins tested. Lysates of HEK293T cells co-expressing full-length MYC–DAPLE (WT or mutants) with the indicated FLAG-tagged G proteins (or no tagged G protein as negative control) were subjected to IP with a FLAG antibody, and bound proteins were detected by IB as indicated. The lower immunoblot panels (Lysates) correspond to aliquots of the starting material used for IPs shown in the upper panels (IP: FLAG). One representative experiment of four is shown for Gαs and Gαq (n = 4), or one representative experiment of two is shown for Gαi3 (n = 2). B, comparison of DAPLE Met-1669 and GIV Val-1679 in the context of their respective Gαi3/GBA motif complex structures. Left panel, homology model of DAPLE GBA motif (green, ribbon representation) in complex with Gαi3 (blue, space-filling representation) was generated using the X-ray crystal structure of the Gαi3/GIV GBA motif complex (PDB code 6MHF). The area of the Gαi3/DAPLE structure model within the dotted box is shown enlarged in the middle panel to illustrate that Met-1669 is largely solvent-exposed. Right panel, detail of the structure of Gαi3 in complex with GIV GBA motif (brown) showing that Val-1669 is also largely solvent-exposed. C, proposed model for the structural basis of DAPLE's G-protein selectivity. Much like GIV Val-1679, DAPLE Met-1669 does not make direct contact with Gαi3. In contrast, DAPLE Met-1669 is required for binding to Gαs or Gαq, suggesting that it makes a contact with these proteins that is not allowed by the shorter chain of the valine located in the corresponding position in GIV.