Figure 2.
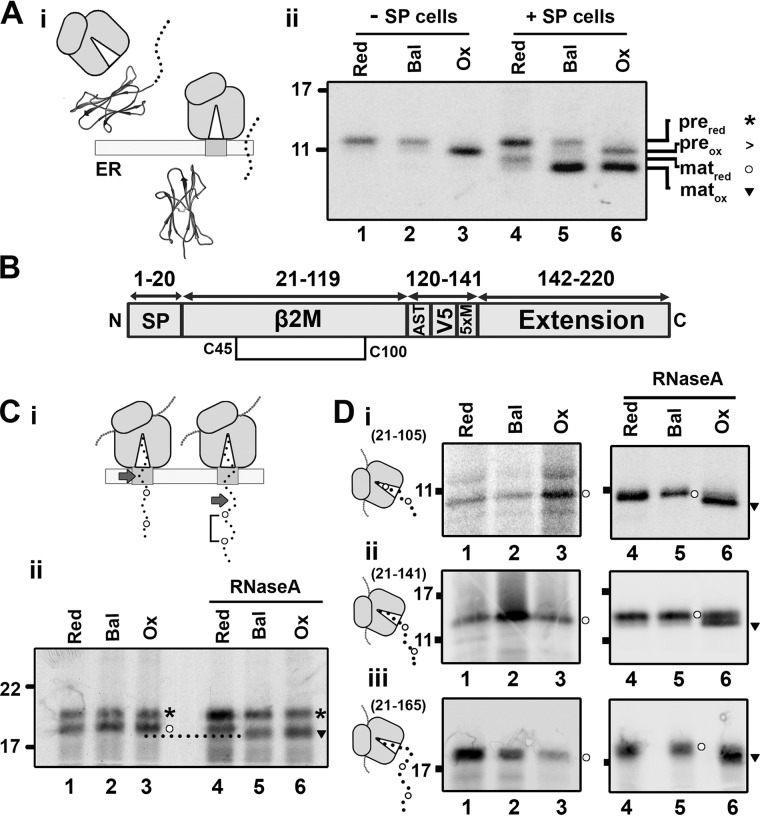
β2M-disulfide formation is folding-driven. A, i, diagram illustrating signal peptide cleavage, which occurs following translocation of β2M. ii, nonreducing SDS-PAGE of immunoisolated WT β2M translated with or without SP cells in reducing (Red), redox-balanced (Bal), or oxidizing (Ox) lysate. The migration of preprotein (pre) and mature protein (mat) when oxidized (ox) or reduced (red) is indicated by the associated symbols (asterisks, >, circles, and inverted triangles). B, organization of the extended β2M construct with the single disulfide (Cys45-Cys100) displayed. C, i, diagram illustrating the dependence of disulfide formation on domain exposure; arrows highlight the end of the β2M folding domain. ii, nonreducing SDS-PAGE shows isolated 175 intermediates translated with SP cells in redox-balanced, oxidizing, and reducing lysate. RNaseA treatment following translation is indicated. D, nonreducing SDS-PAGE analysis of isolated ribosome/nascent chain complexes representing β2M 21–105 (i), 21–141 (ii), and 21–165 (iii) translated in redox-balanced, oxidizing, or reducing lysate without SP cells. Samples treated with RNaseA following translation are indicated. Schematics next to each panel indicate the predicted cysteine exposure at each intermediate length. All gels in this figure are representative of at least three repeats.