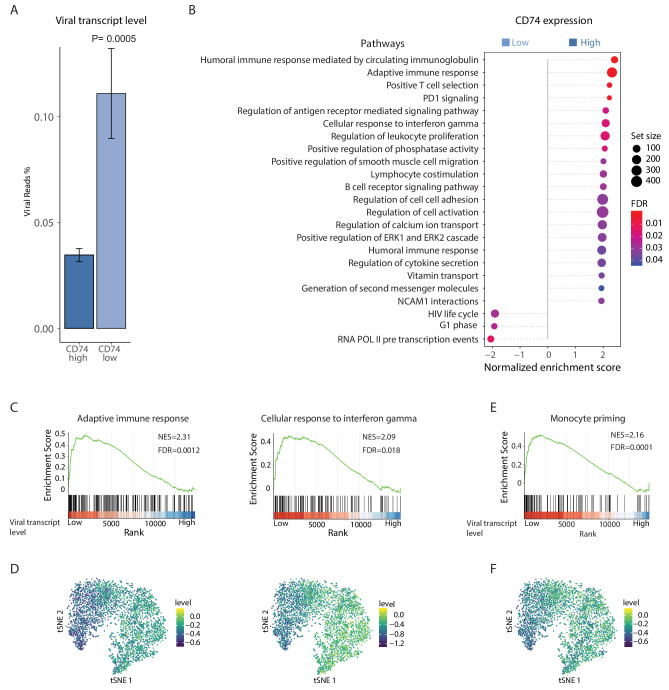
Figure 6. HCMV latency in monocytes is associated with reduced immune-response gene signatures.
RNA-seq was performed on HCMV- infected CD14+ Monocytes that were sorted according to cell-surface levels of CD74 at 3dpi. (A) Normalized viral gene expression in CD74high and CD74low cells. P value, calculated using likelihood ratio test on logistic regression of viral reads, is indicated. (B) Summary of gene set enrichment analysis (GSEA) of differential expressed genes identified in RNA-seq analysis of CD74high and CD74low cells using annotated GO biological processes and Reactome pathways. (C) Representative pathways from GSEA of genes ranked by their differential expression between CD74high and CD74low cells. (D) tSNE plot of scRNA-seq of latent monocytes colored by expression level of the pathways shown in C. (E) Monocyte priming gene set from Velten et al. (2017) analyzed on GSEA. Genes are ranked by their differential expression between CD74high and CD74low monocytes. (F) tSNE plot of scRNA-seq of latent monocytes colored by the expression level of the monocyte priming gene set from Velten et al. (2017).
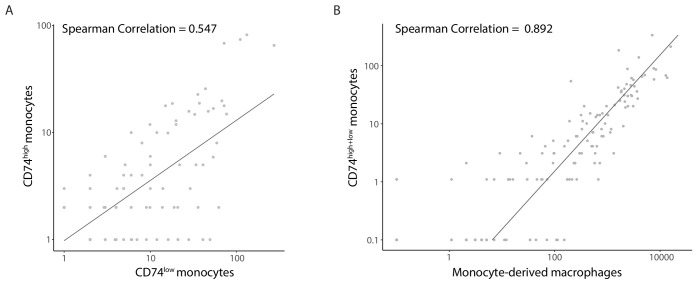
Figure 6—figure supplement 1. Viral gene expression profile of infected CD14+ monocytes correlates between CD74high and CD74low cells and to late lytic profile.
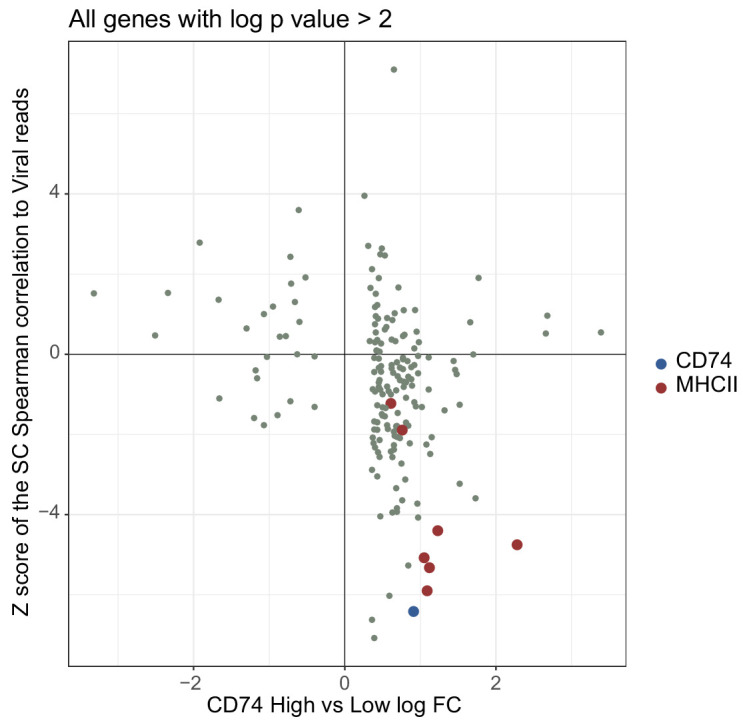
Figure 6—figure supplement 2. Comparison of changes detected in bulk RNA-seq and scRNA-seq data.