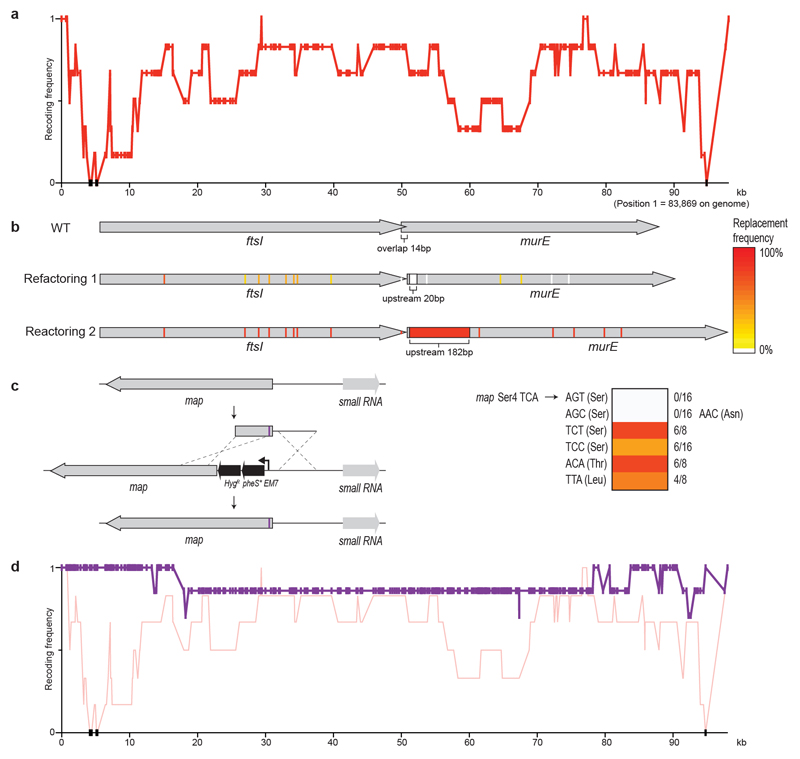
Extended Data Fig. 2. Recoding ftsI-murE and map in fragment 1.
a, Recoding landscape of fragment 1. We sequenced six clones after REXER. Each dot represents the frequency of recoding within the sequenced clones (y axis) for a target codon at the indicated position in the genome (x axis). Black dots indicate positions at which we did not observe recoding. Four codons and a refactoring of ftsI-murE, and one codon in map, were rejected.
b, Refactoring the 14-bp overlap of ftsI and murE. The codons and overlaps are colour-coded by their post-REXER replacement frequency in the clones sequenced. Using our initial refactoring scheme (refactoring 1) (in which the overlap plus 20 bp of upstream sequence was duplicated), we did not observe replacement of the overlap by synthetic DNA (in the six clones sequenced after REXER). Refactoring scheme 2 (refactoring 2) (which duplicates the overlap plus 182 bp of upstream sequence) resulted in complete recoding of this region in 12 of the 16 post-REXER clones that we sequenced.
c, Testing alternative codons at Ser4 in map. A double-selection cassette, pheS*-HygR, on a constitutive EM7 promoter was introduced upstream of map, followed by a ribosome-binding site. We replaced the cassette using linear double-stranded DNA that introduces alternative codons (purple bar) at position four, via lambda-red recombination and negative selection for loss of pheS*. DNA with AGC and AGT did not integrate (0/16 clones); we recovered one clone for AGC but sequencing revealed that it contained a mutant AAC (Asn) codon. TCT (6/8), TCC (6/16), ACA (6/8) and TTA (4/8) were allowed.
d, Recoding landscape (purple) over the genomic region shown in a, following REXER with a BAC that contained refactoring scheme 2 for the ftsI-murE overlap and TCT at position 4 in map. In total, 2/7 post-REXER clones were completely refactored and recoded, and each target codon was replaced in at least 5/7 clones. The data from a are shown in red for comparison.