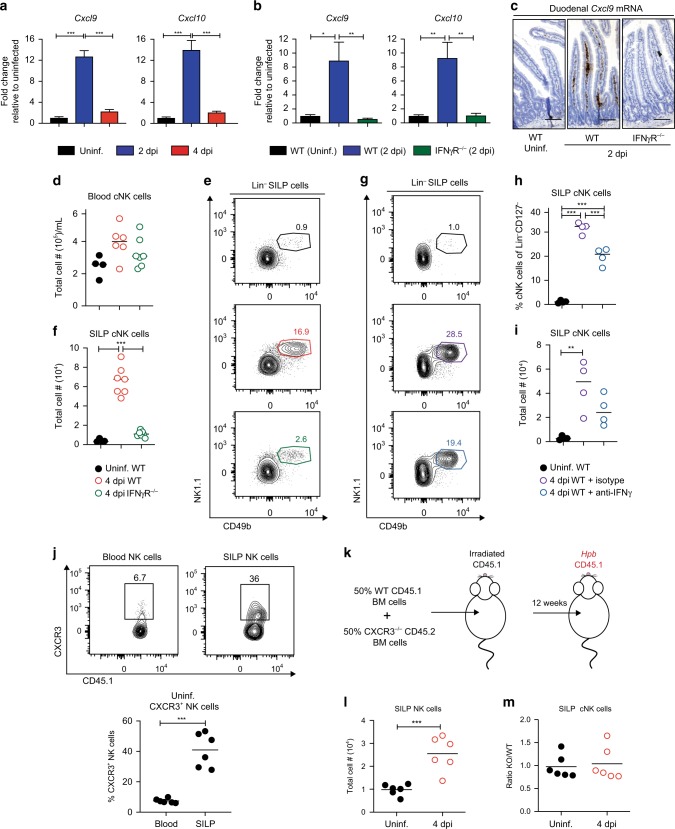
Fig. 5.
IFNγ-dependent, CXCR3-independent recruitment of NK cells to the SILP during Hpb infection. a Fold change in Cxcl9 and Cxcl10 mRNA expression in WT duodenal tissue of WT mice relative to uninfected WT duodenal tissue at 2 and 4 dpi (n = 7–10). b Fold change in Cxcl9 and Cxcl10 mRNA expression in WT and IFNγR−/− duodenal tissue relative to WT uninfected duodenal tissue at 2 dpi (n = 4–7). c RNA Scope for Cxcl9 transcripts, in brown, in the duodenal tissue of uninfected WT and 2 dpi of WT and IFNγR−/− mice (scale bar, 100 μm). d Total numbers per mL of blood NK cells (CD127−NK1.1+CD49b+) in uninfected WT or 4 dpi of WT and IFNγR−/− mice. e Representative contour plots and f total numbers of cNK cells in the SILP of uninfected WT or 4 dpi of WT and IFNγR−/− mice. g Representative contour plots, h frequency, and i total numbers of cNK cells in the SILP of uninfected mice, or 4 dpi of mice treated with anti-IFNγ-blocking antibody or isotype control. j Representative contour plots and frequencies of CD127−NK1.1+CXCR3+ NK cells in the blood and SILP of uninfected WT mice. k BM chimera experimental design. l Total cell number of CD127−NK1.1+ NK cells in uninfected and 4 dpi BM chimera mice. m Ratio of KO/WT SILP cNK cells in uninfected and 4 dpi of BM chimera mice. Data shown are pooled from three a and two b, d–f, j, l, m independent experiments and representative of two independent experiments g–i. a, b, d, f, h, i Data were analyzed by one-way ANOVA with Tukey’s post test for multiple comparisons. j, l, m Data were analyzed using an unpaired parametric t test (*p < 0.05, **p < 0.01, ***p < 0.001). Each dot represents an individual mouse. Error bars, SEM.