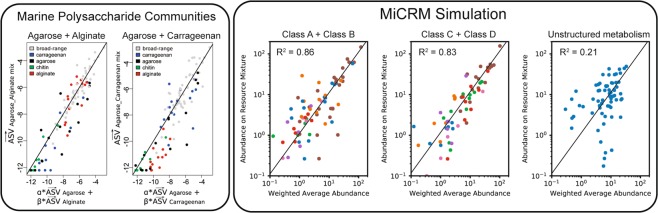
Figure 10.
Modularity of community assembly. Left: In the experiments reported in21, synthetic beads composed of different kinds of polysaccharides, including agarose, alginate and carrageenan, were incubated with coastal seawater and colonized by the marine bacteria resident in the seawater sample. 16S rRNA amplicon profiling was performed for communities grown on beads composed of a single kind of polysaccharide, as well as mixtures of two kinds of polysaccharides. Relative abundances of amplicon sequences variants for two different mixtures (Agarose/Alginate and Agaraose/Carrageenan) are plotted versus a weighted average of the relative abundances on the pure beads. Solid lines are fits to a linear mixture model, with R2 of 0.84 and 0.74, respectively. Right: Abundance of each species in simulated communities supplied with mixtures of two resource types, plotted against the average of the abundances for communities supplied with just one of the resource types, with the total energy supply held constant. For the first two panels, all other parameters are the same as for the human microbiome simulations of Fig. 6, except that each sample is initialized with all 5,000 species from the regional pool. Titles indicate the class labels of the two supplied resources for each scenario, and species are colored by metabolic family following Fig. 4. In the third panel, simulations were run with the same number of resources and species, but with all resources assigned to the same resource class, eliminating all metabolic and taxonomic structure. Solid lines are predictions of the additive model where the abundance in the mixture equals the average of the abundances in the single-resource condition. The R2 score of this model is also shown in each panel.