Abstract
Aedes aegypti is the primary vector for transmission of Dengue, Zika and chikungunya viruses. Previously it was shown that Dengue virus infection of the mosquito led to an in increased expression of the odorant binding protein 22 (AeOBP22) within the mosquito salivary gland and that siRNA mediated knockdown of AeOBP22 led to reduced mosquito feeding behaviors. Insect OBPs are implicated in the perception, storage and transport of chemosensory signaling molecules including air-borne odorants and pheromones. AeOBP22 is unusual as it is additionally expressed in multiple tissues, including the antenna, the male reproductive glands and is transferred to females during reproduction, indicating multiple roles in the mosquito life cycle. However, it is unclear what role it plays in these tissues and what ligands it interacts with. Here we present solution and X-ray crystallographic studies that indicate a potential role of AeOBP22 binding to fatty acids, and that the specificity for longer chain fatty acids is regulated by a conformational change in the C-terminal tail that leads to creation of an enlarged binding cavity that enhances binding affinity. This study sheds light onto the native ligands for AeOBP22 and provides insight into its potential functions in different tissues.
Subject terms: X-ray crystallography, Solution-state NMR, Carrier proteins, Fatty acids
Introduction
A critical step in disease transmission by hematophagous mosquitoes is the location of a human host for a blood meal by the female mosquito. Host location and selection of biting sites is driven by the perception of chemosensory stimuli that requires the interplay of a number of factors including chemosensory receptors and odorant binding proteins (OBPs)1,2. Ae. aegypti OBP22 (AeOBP22) is a member of the OBP family of proteins that has been directly implicated in regulating these feeding behaviors3. AeOBP22 is unusual in that it is expressed in multiple chemosensory tissues, including in the antenna, the proboscis of the females, the thoracic spiracles4, in the male reproductive glands where it is transferred to the females during mating4,5, and in the salivary glands3,6,7. Surprisingly, it was discovered that, in combination with other chemosensory genes, its expression in the salivary glands is up regulated in response to Dengue virus (DENV) infection and that knockdown of AeOBP22 using dsRNA approaches led to reduced blood feeding behaviors3
OBPs were first identified through their role as pheromone binding proteins (PBPs), which are components of the chemosensory apparatus that are secreted into the lymph fluid surrounding the neuronal dendrites of the chemosensory sensilla8. PBPs are a subgroup of OBPs that bind preferentially to pheromones. OBPs are essential for many aspects of chemosensory signal transduction9,10. In the lymph it is proposed that OBPs function to transport hydrophobic ligands across the aqueous lymph fluid and deliver them to chemosensory receptors1,2,11,12. Many studies have provided support for this hypothesis, particularly for perception of pheromonal compounds10,13–17. However, other studies have suggested that lymph fluid is in fact an emulsion of fatty acids and that these can enhance dispersion of pheromones into the lymph and promote interactions with the OBP18. Additional evidence suggests that OBPs may rather function to sequester ligands to regulate the gain/sensitivity of the chemosensory response19,20.
Increasingly it is apparent that OBPs are also expressed in outside of the primary chemosensory tissues including in hemolymph21, reproductive tissue4,5,22–24, as components of mosquito eggshells23,25–27, and as secreted components of the salivary glands of multiple insects28,29. In particular it has been proposed that the D7 family of OBP related proteins in mosquito saliva function to limit inflammation30–32, and blood clotting through their ability to sequester pro-inflammatory signals including biogenic amines and cysteinyl-leukotrienes33. The expression of AeOBP22 in the antenna and proboscis clearly implicates it in regulating responses to host-derived odors that emanate from skin and/or sweat that drive blood feeding behaviors, while its expression in the salivary gland suggests the potential for it to be transferred to the human host during a blood meal. It is well established that components of salivary gland extracts (SGEs) dramatically impact blood feeding behaviors and viral infectivity33–36. In order to better understand the potential roles of AeOBP22, we have undertaken a structural and biophysical characterization of the protein with the aim of understanding its ligand binding properties. These studies suggest that AeOBP22 may have evolved to bind to a range of fatty acids and that binding selectivity for longer chain fatty acids (>12 carbon atoms) is achieved through a conformational change in the C-terminal tail that leads to the formation of an expanded ligand binding pocket.
Results
NMR spectroscopy identifies long chain fatty acids as ligands for AeOBP22
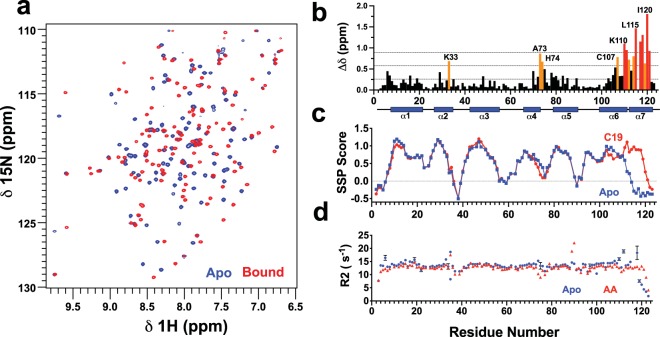
We used NMR spectroscopy to screen compounds from human sweat and skin, repellents, fatty acids and bioactive lipids for binding to AeOBP22. We prepared purified, delipidated protein37–39 by extensively washing isolated inclusion bodies using buffer containing 1 M urea followed by refolding40. Retrospectively, we determined that “non-delipidated” samples contain a mixture of the apo-state of the protein and the complex formed with palmitic acid (16 carbons) (Supplementary Fig. 1). The 1H-15N HSQC spectrum of the fully delipidated sample (Fig. 1a) shows 119 of the expected 120 peaks for the backbone amides. In NMR screening, we consistently found that longer chain fatty acids (C16–C20 carbons) produce large changes in the appearance of the NMR spectrum at low concentrations (Fig. 1a). Other compounds can produce the same magnitude of NMR chemical shift changes, notably geraniol, citronellol and benzaldehyde (Supplementary Fig. 2). However, this only occurred at high ligand concentrations (typically> 1 mM) and binding is weak as exemplified by the concentration dependence of the chemical shift changes. In contrast, fatty acids bind with high affinity as evidenced by the presence of peaks from the free and the bound states present in slow exchange on the NMR timescale when the ligand is present in sub-stoichiometric quantities (Supplementary Fig. 3). From the NMR chemical shift assignments of the apo-protein and the complexes with nonadecanoic (C19) acid and arachidonic acid (AA)40 we determined that fatty acid binding has the largest impact on residues 106–121 in the C-terminus (Fig. 1b). Further calculations of the secondary structural propensities (SSPs) from the chemical shift data41 predict that the C-terminal tail adopts an extended conformation in the apo-state and adopts an α-helical conformation when bound to longer chain fatty acids (Fig. 1c)40.
Figure 1.
Conformational changes on binding of fatty acids to AeOBP22 (a) Region of the 1H-15H HSQC of apo-AeOBP22 (blue) and bound to nonadecanoic acid (red). (b) Plot of normalized chemical shift changes between apo-AeOBP22 and bound to C19 fatty acid. Significant chemical shift changes are color coded as greater than 1 s.d. above the mean (orange) and 2 s.d. above the mean (red). Horizontal dashed lines indicate the position of the mean, +1.sd. and +2 s.d. from bottom to top. The location of the α-helical regions is shown as blue cylinders below. (c) Plot of secondary structure propensity scores (SSP)41 for apo AeOBP22 (blue) and the complex with C19 (red). (d) Plot of NMR 15N R2 relaxation rates for apo (blue) and bound to arachidonic acid (red) recorded at a 1H frequency of 900 MHz. Error bars are shown in black.
Crystal structures of AeOBP22 complexes
Next we used X-ray crystallography to better define the conformational changes that occur on binding fatty acids and we determined structures in three different crystal forms. Initial crystals formed in the P3121 space group, and the structure was solved using single wavelength anomalous dispersion (SAD) of a tantalum bromide (Ta6Br12) soaked crystal refined at 2.6 Å resolution. A native data set collected on the same crystal was refined at a resolution of 1.9 Å to an R/RFree of 18.3/20.1% (Supplementary Fig. 4a). In this crystal form the protein forms a domain swapped dimer, with residues 116–122 of one molecule forming an anti-parallel β-sheet with residues 37–41 in a symmetry related molecule (Supplementary Fig. 4b).
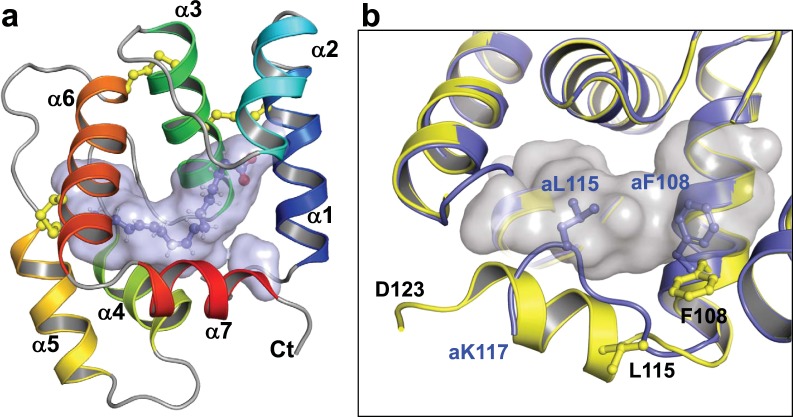
The second crystal form was solved in the P31 space group and contains nine molecules in the asymmetric unit arranged in a pseudo three-fold arrangement of “trimers” consisting of domain swapped dimer and a separately packed monomer (Supplementary Fig. 5). The dimers are identical to those observed in crystal form 1 and superimpose with an average pairwise RMSD of 0.47 Å. The individual monomers superimpose to each other with an average pairwise RMSD of ~0.2 Å. Ligands are observed bound at the center of each monomer (Fig. 2a and 3a) with residues 112–121 forming an α-helix that forms one edge of the ligand-binding pocket (Fig. 2b). No ligand is observed in the dimeric components. In this form we solved the structures with palmitic acid (C16:0), palmitoleic acid (C16:1), and eicosanoic acid (C20) complexes.
Figure 2.
Structure of AeOBP22. (a) Ribbon diagram of the AeOBP22-linoleic acid complex solved by X-ray crystallography at 1.85 Å. Helices are color coded from blue to red, N to C terminus. The binding pocket is shown as a surface representation and linoleic acid is shown as sticks. (b) Comparison of the C-terminal tail in the linoleic acid complex (yellow) and in the apo-state (blue). In apo AeOBP22, L115 and F108 insert into the pocket (shown in grey) to occlude binding of larger ligands. Reside numbers for the apo-state are preceded with an “a”. Residues 118–123 of the apo-state are not observable in the crystal.
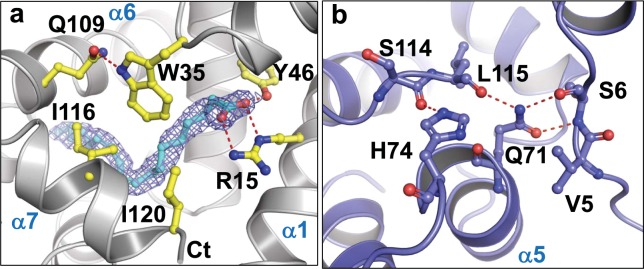
Figure 3.
Critical interactions that stabilize the in the bound and apo-states of AeOBP22 (a) Electron density for linoleic acid in a 2Fo-Fc omit map contoured at 1σ with the fatty acid modelled in cyan. Y46 and R15 make specific H-bonds with the fatty acid. Positioning of the head group is reinforced by interactions with I120, I116, and W35, which H-bonds to Q109 in helix-6. (b) In apo-AeOBP22, the position of the C-terminal tail is stabilized by H-bonds between S114 and L115 with the side chains of H74 in the α4-α5 loop and Q71 in helix-4, which in turn forms hydrogen bonds with S6 in the N-terminal region.
The third crystal form grows in the presence of cadmium and/or cobalt ions in the C121 space group and contains only monomers. Structures of the apo-state and the complexes formed with linoleic acid (C18:2) and AA (C20:4) were solved using cobalt SAD methods and refined to a resolution of 1.85 Å. Data collection and refinement statistics for deposited structures are given in Supplementary Table 1.
AeOBP22 is monomeric in solution
Previous studies have proposed that dimerization of OBPs may regulate ligand binding and transport19,42–47. Therefore, we used measurements of 15N NMR relaxation rates, R1 and R2, to determine if AeOBP22 forms dimers or higher order complexes. For a globular protein there is a direct correlation between the average R2/R1 ratio and the molecular weight48–51. For AeOBP22 the predicted molecular weights from multiple measurements of the R2/R1 ratios in both apo and fatty acid bound states (Supplementary Fig. 6) were in the range 14.1–15.2 KDa, in agreement with the expected molecular weight of 14.3 KDa, indicating that the protein exists as monomers in the absence of other binding partners. We saw no evidence of any significant dimerization over long periods even at the high concentrations used for NMR chemical shift assignments (600–700 μM). Therefore, we conclude that the domain swapped dimers are an artifact of the crystallization process, and further discussions below are confined to the structures of the monomeric forms.
A complete analysis of the relaxation data using Relax52–54 (Supplementary Fig. 7) revealed slightly elevated exchange contributions to the relaxation rates for residues 111–118 suggesting that this region of the protein may undergo conformational exchange on slower time scales, and this may be important to allow access of fatty acids to the ligand binding pocket. This analysis also confirms that residues 74–78 exhibit increased conformational flexibility in the bound state compared to the apo-state.
Description of structure
The monomeric forms of AeOBP22 are similar to other classical OBPs which consist of six α-helices stabilized by three disulfide bridges surrounding a hydrophobic pocket (Fig. 2a). In the bound state, AeOBP22 is unusual in that it contains a seventh C-terminal α-helix that forms one edge of the ligand-binding pocket. The ligand-bound monomers from all our structures superimpose well with a pairwise RMSD of 0.40 Å (Supplementary Fig. 8). Small differences are seen in the position of helix-7, which rotates out slightly from the binding pocket in the presence of larger fatty acids such that the Cα at Ile120 is approximately 2 Å displaced from its position with shorter chain fatty acids. Additionally, we see variations in the position of the α4-α5 loop (residues 73–78), which may be a result of crystal packing interactions.
The ligand-binding pocket is in the form of a long tunnel approximately 20 Å in length and occupies 144 ± 10 Å3 calculated using CASTp 3.055, and the alkyl chain of the ligand contacts hydrophobic residues that line the pocket (Supplementary Fig. 9). At the opening of the pocket there is an electrostatic patch formed by Arg15, Lys33 and Lys117 (Supplementary Fig. 10). Arg15 in combination with Tyr46 make multiple specific H-bonds to the carboxyl group of the fatty acid (Fig. 3a) and together define the requirement for a negatively charged group at this position. The fatty acid head group also contacts Trp35, which in turn hydrogen bonds to Gln109. All these residues are highly sensitive to ligand binding in NMR experiments.
At the distal end of the pocket from Arg15, three highly ordered water molecules hydrogen bond to Trp100, Ala101, Gly104, Cys88 and Val89. Analogous water molecules are observed in the binding pocket of Ae. aegypti Juvenile Hormone Binding Protein (AeJHBP)56. This suggests that AeOBP22 may accommodate ligands with polar groups at this position. To test this, we examined the interactions with 16-hydroxy-hexadecanoic acid (C16-OH) and observed large NMR chemical shift changes relative to the apo-protein that are comparable to the changes observed with C16 (not shown). We observed additional changes for residues 86–92 that are consistent with the interaction of the C16-hydroxyl group at this position (Supplementary Fig. 11). However, residues throughout helices 6 and 7 (103–121) show line broadening and reduced intensity indicative of increased conformational averaging. Additionally, we found that C16-OH does not compete for binding of a fluorescent reporter in ligand binding assays (below) and so we conclude that distal polar groups appear to be unfavorable for binding and destabilize the conformation of the AeOBP22 complex, suggesting that the role of the buried water molecules is more likely to be as a structural component.
Structure of the apo-state of AeOBP222
The structure of apo-AeOBP22 confirms that residues 118–123 are disordered and 112–117 adopt an extended structure compared to the α-helix observed in the bound state (Fig. 2b and Supplementary Fig. 8b). In this extended conformation, Leu115 and Phe108 insert into the core of the protein and restrict the size of the binding pocket (Fig. 2b). This extended structure is stabilized by the formation of hydrogen bonds between Ser114 and Leu115, with His74 and Gln71 in the α4-α5 loop respectively (Fig. 3b). In turn, Gln71 hydrogen bonds with Ser6 in the N-terminus. This network of interactions links conformational changes in the C-terminus to the N-terminus through the α4-α5 loop. This explains our NMR relaxation data (Fig. 1d and Supplementary Fig. 6 and 7), which shows increased conformational mobility of the α4-α5 loop on binding fatty acids because these interactions are disrupted.
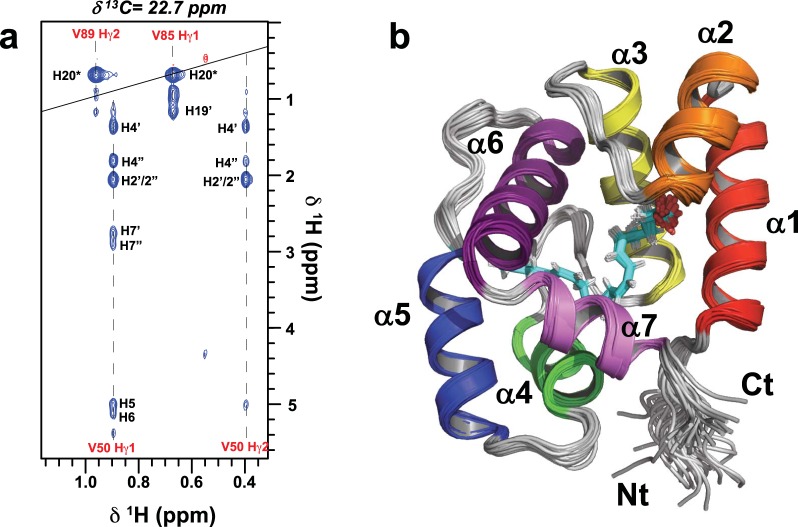
NMR Solution Structure of AeOBP22 with Arachidonic Acid
In parallel with our crystallographic studies we determined the NMR solution structure of the AeOBP22-AA complex. AA was chosen because the vinylic protons of this fatty acid each have a unique chemical shift and are in a region of the spectrum (4.6–5.6 ppm in 1H) that has minimal overlap with resonances from the protein40. This increased chemical shift dispersion greatly facilitated the assignment of intermolecular NOEs between the protein and the ligand (Fig. 4a). The ensemble of 30 lowest energy structures from the final round of calculations superimpose with a backbone RMSD of 0.32 Å from the mean structure, while the carbon atoms in the ligand superimpose with an overall RMSD of 0.31 Å (Fig. 4b). The list of structural restraints and refinement statistics are given in Supplementary Table 2.
Figure 4.
NMR Structure of the AeOBP22-AA Complex (a) A slice through a 12C-edited/13C-filtered intermolecular NOESY spectrum at δ13C = 22.7 ppm showing NOEs between resonances from the protein (labeled in red) and arachidonic acid (black labels). For the lipid the hydrogens are numbered according to the attached carbon in the alkyl chain. (b) Superposition of the 30 lowest energy structures from the final iteration of NMR calculations (RMSD = 0.32 Å for res 7–121), helices 1–7 are color coded from red-violet. The arachidonic acid is shown in cyan.
The NMR structure of the AeOBP22-AA complex superimposes with the crystal structure with an RMSD of 0.67 Å for the backbone atoms of residues 7–120 and confirms that the monomeric structure observed in the crystal is maintained in solution. The biggest differences between the structure are in the conformation of the loop between helices 4 and 5, which are involved in crystal packing, but which are dynamic in solution. The overall structure of the arachidonic acid ligand is similar to the structure observed in the crystal. However, even in the crystal there are differences in the ligand position in the two monomers in the asymmetric unit.
Impact of Chain Length on the Conformation of the C-terminal tail
In the apo-AeOBP22 structure we observed a small binding pocket located between Arg15 and Leu115 that could potentially accommodate short chain fatty acids (up to C6). When we examined the binding of fatty acids containing up to 8 carbons by NMR, we observed chain length dependent chemical shift changes for residues in the N-terminal half of the protein (Fig. 5a and Supplementary Fig. 12a), that are consistent with binding of the carboxylate group to the basic residues in the vicinity of Arg15, Lys33 and Trp35 (Supplementary Fig. 12b). To ensure that addition of free fatty acids did not result in changes in pH that could perturb the NMR spectrum, we examined chemical shift changes caused by pH alone (Supplementary Fig. 12b). We observed that the patterns and magnitude of chemical shift changes caused by short chain fatty acids are distinct from those caused by changes in pH alone. It was surprising that C8 still binds with relatively little impact on the overall structure suggesting a significant plasticity in the binding pocket. However, the exact mode of binding of short chain fatty acids remains to be definitively established.
Figure 5.
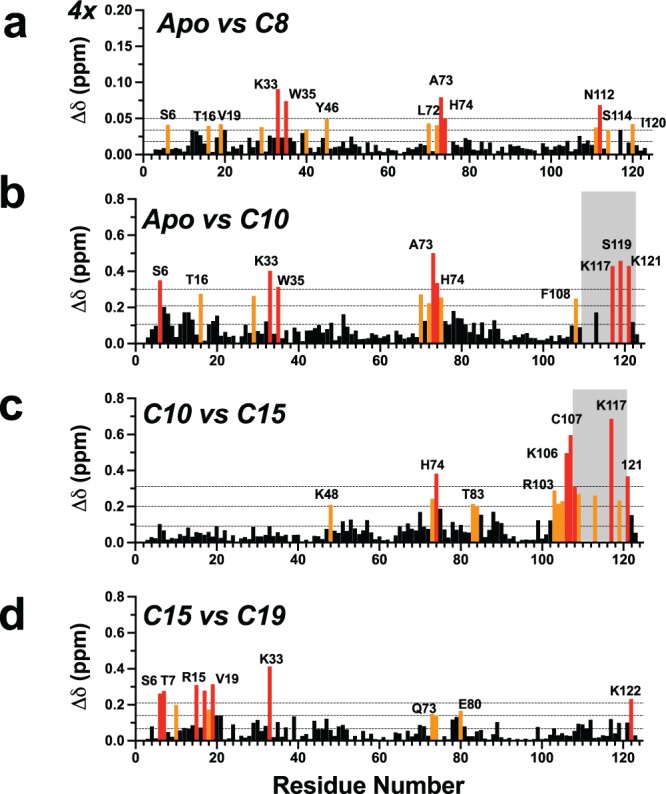
Chemical shift changes as a function of fatty acid chain length. Comparison of chemical shift differences in the 1H-15N HSQC spectra recorded at 600 MHz for (a) apo compared to octanoic acid. (b) Apo versus C10. (b) C10 acid vs C15 acid and (d) C15 vs C19 acids. Significant chemical shift perturbations are color coded as in Fig. 1. In panel (a), the vertical scale has been expanded by four compared to the other panels. For C10–C14 conformational averaging in the C-terminal tail limited the assignments for a number of residues in this region (shaded in grey in b and c). In all cases the protein was at ~100 μM and the fatty acid was present at a concentration of 200 μM in sodium phosphate at pH 6.5.
In contrast to short chain fatty acids, large chemical shift perturbations are observed throughout the protein in the presence of decanoic acid (C10) (Fig. 5b). After reassigning the spectrum, we determined that C10 binding impacts the C-terminal tail, the α4-α5 loop, the end of helix-2 (residues 33–35) and helix-1. Several residues in the C-terminal tail show increased exchange broadening and could not be assigned (shaded in grey in Fig. 5b,c). Increasing the alkyl chain from C10 to C15, leads to additional perturbations in the C-terminus (aa 109–121) and in the α4-α5 loop (aa 70–75). However, when the alkyl chain is further increased from C15 to C19 additional chemical-shift perturbations localize predominantly to residues 6–21 in α-helix 1 (Fig. 5d), with limited changes in the C-terminal region. There are no additional changes beyond C20, and fatty acids with more than 20 carbon atoms show a dramatic reduction in their ability to bind (not shown). We conclude that fatty acids with at least 10 carbon atoms are required to stimulate a conformational change in the C-terminus, and this is maximally induced with a chain length of 15–16 carbon atoms. Secondly, a conformational shift in helix-1 is required to accommodate binding of longer chain fatty acids C17–C20. This adaptation differs to that observed in the crystal, where helix-7 shifts in response to longer chain fatty acid. We attribute this to the crystal packing contacts formed by helix-1 which restricts its ability to move. In contrast, helix-7 makes few crystal contacts and has greater ability to adapt its conformation to accommodate different ligands.
Fatty acid chain length and degree of unsaturation impact the binding affinity to AeOBP22
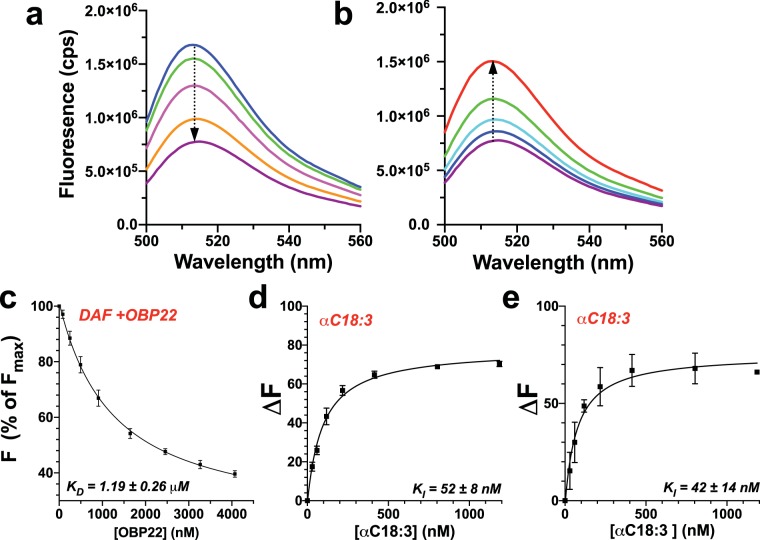
Next, we asked how the chain length and presence of unsaturation impacts the binding affinity for AeOBP22. Many previous studies have used 1-NPN as a fluorescent reporter to examine ligand binding to OBPs57,58, including for AeOBP224,59. In our hands, we found that 1-NPN was unsuitable for studies of fatty acid binding as it bound with low affinity and showed non-specific interactions that confounded analysis. Further, studies with 1-NPN are often confounded by the ability of ligands to bind simultaneously and quench 1-NPN fluorescence rather than bind in a competitive manner60. Therefore, we investigated the use of a fluorescently labeled fatty acid derivative 5-(N-dodecanoyl)-amino-fluorescein (DAF), as a reporter for competition binding assays. DAF shows a strong fluorescence emission with a maximum at 513 nm which is quenched upon addition of AeOBP22 (~75% reduction from maximal intensity) and with a KD of 1.19 ± 0.26 μM (n = 7) (Fig. 6a,c). Addition of fatty acids with 14 or more carbon atoms releases DAF from AeOBP22 leading to a recovery of the initial fluorescence (Fig. 6b,d). In addition, NMR spectroscopy shows that DAF produces chemical shift changes comparable to those produced by C11–C14 fatty acids (Supplementary Fig. 13), and conformational line broadening for residues in the C-terminal region. Therefore, we conclude that the alkyl chain of DAF likely binds to the central pocket of AeOBPP2 and can displace the C-terminal tail, and that fatty acids bind competitively with DAF.
Figure 6.
Long chain fatty acids bind with high affinity to AeOBP22. (a) Florescence emission spectra of DAF (blue) excited at 490 nm, with increasing amounts of AeOBP22 (arrow) shows concentration dependent quenching. (b) Titration of the final sample in a, with increasing linoleic acid (arrow) shows that the fatty acid competes for binding of DAF and recovery of initial fluorescence (red). (c) Determination of binding constant for AeOBP22 and DAF from 7 replicates of experiments shown in panel (a). DAF concentration was 84 nM. Plot of the intensity at the florescence maximum (513 nm) presented as change from initial fluorescence. Data were fit according to Eq. 2. (d) Determination of the binding constant of α-linoleic acid at pH 8.0 from multiple replicates (n = 3) of experiments shown in panel (b) by fit of the raw intensity using Eqs. 2 and 3 using a KD for DAF of 1.191 μM. The DAF concentration was fixed at 100 nM and the protein at 1000 nM. Data are presented as normalized recovery of the quenched fluorescence for ease of viewing. (e) The same experiment as in (d) but recorded at pH 5 (n = 2). Intrinsic DAF fluorescence is significantly quenched at pH 5.0 leading to increased error in measured points.
We used DAF based competition binding assays to determine binding affinities for a series of fatty acids (Table 1 and Supplementary Fig. 14). This revealed that the both chain length an unsaturation impact the binding affinity. Saturated C16 binds weakly with KD ~690 nM, however, introduction of a single degree of unsaturation at the Δ9 position (C16:1) leads to an approximate four-fold increase in the binding affinity (KD = 175 ± 41 nM). Increasing the chain length and the introduction of additional unsaturation enhances binding, which appears to be optimal for the fatty acids containing 18 carbon atoms, as increasing the chain length to C20 leads to a two-fold reduction in affinity. However, the extent of unsaturation also impacts the affinity, with the more rigid AA (C20:4) having a lower affinity than the monounsaturated C20:1. These results are consistent with our NMR data that shows binding of longer chain fatty acids requires a conformational change in α-helix 1 (Fig. 5d) and so this appears to be unfavorable for binding.
Table 1.
Dissociation constants for binding of fatty acids to AeOBP22.
| Ligand | KD (nM)1 |
|---|---|
| DAF | 1191 ± 2612 |
| C16:0 | 689 ± 89 |
| C16:1 | 175 ± 41 |
| C18:1 | 84 ± 5 |
| C18:2 | 104 ± 30 |
| αC18:3 pH 8.0 | 52 ± 8 |
| pH 5.0 | 42 ± 143 |
| γC18:3 | 111 ± 27 |
| C20:1 | 102 ± 16 |
| C20:4 | 286 ± 29 |
1Values reported in nM and ± the standard deviation, n = 3.
2n = 7.
3n = 2.
Changes in pH do not impact the binding of fatty acids
In apo-AeOBP22, His74 makes interations that maintain the C-terminal residues in a conformation that occludes the ligand binding site. Consequently it is possible that pH may impact the conformation of the C-terminal residues and ligand binding in a manner somewhat analogous to that observed for PBPs61–64. Indeed, previous studies of AeOBP22 have suggested that the helical content of the protein increases at lower pH59. To test this, we compared the binding affinity for α-linoleic acid at pH 6.5 and 5.0 but found no significant difference at these two pHs, 52 ± 8 nM vs 42 ± 14 nM respectively (Fig. 6d,e and Table 1). We could not use DAF below pH 5.0 because its intrinsic fluorescence is severely quenched. Therefore, we used NMR to examine the effect of pH on the protein structure and binding. For these studies, samples were prepared in sodium citrate at pH 6.5 and the pH adjusted using hydrochloric acid over the range 6.5–4.5. We found that lowering the pH resulted in chemical shift changes consistent with protonation of His74 and His70 between pH 6.5 and 5.5 (Supplementary Fig. 14b) and sidechain carboxyls between pH 5.5 and 4.5 (not shown). However, the cumulative chemical shift changes (~0.3 ppm) over the pH range 6.5–4.5 are 5–6 times smaller than the perturbations induced by the binding of chain fatty acids (Fig. 1), indicating that changes in pH do not induce the conformational rearrangement observed with fatty acids. To validate this, we obtained backbone chemical shift assignments (1H, 15N, 13Cα, 13Cβ) for both the apo-protein and the α-linoleic acid complex at pH 4.5 and examined differences in the secondary structural propensities and the normalized chemical shift differences between the two samples (Supplementary Fig. 15). These almost exactly matched the SSPs and patterns of chemical shift changes upon ligand binding observed for samples recorded at pH 6.5, and confirms that pH does not impact the structure of the apo-protein or the ability of AeOBP22 to bind to longer chain fatty acids at low pH.
Comparison of AeOBP22 with known structures
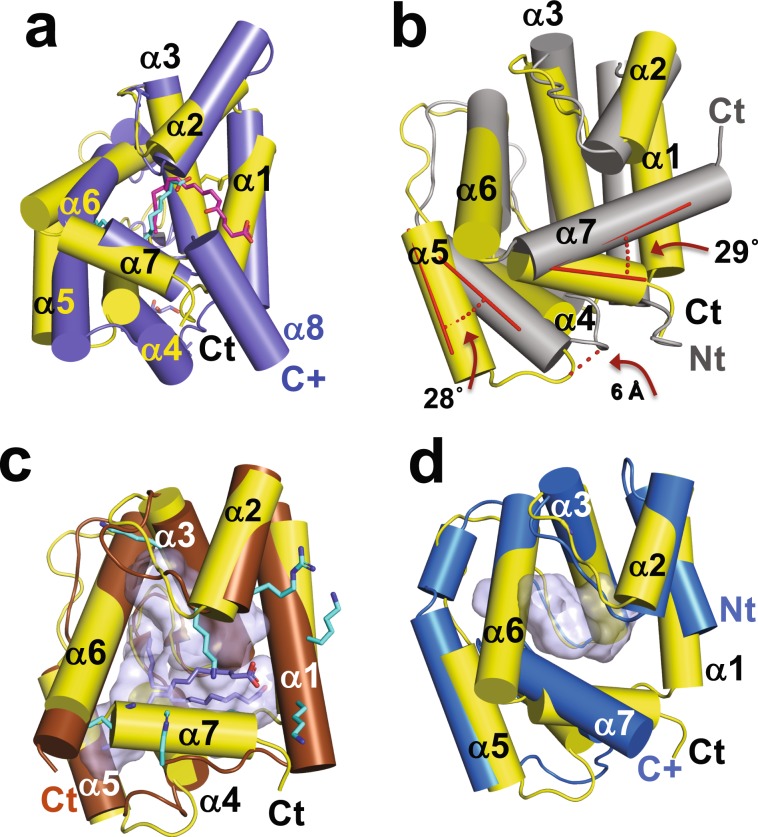
The bound state of AeOBP22 contains a seventh α-helix at the C terminus that forms one edge of the ligand-binding pocket. A DALI search65,66 shows the highest structural similarity to the N-terminal domains of the insect D7 proteins that contain dual OBP-domains; Ae. aegypti juvenile hormone binding protein56 (AeJHBP, PDB-ID 5V13) with a DALI Z-score of 15.4, An. stephensi D767 (AsteD7, PDB 3NHT, Dali Z = 12.8), and An. gambiae D7-Leukotriene E4 complex (AgamD7, PDB 3DZT, Dali Z = 12.2)31. The seventh helix of AeOBP22 is in a remarkably similar position to the seventh helix of these D7 proteins (Fig. 7a, only the N-terminal domain of AsteD7 is shown). However, in the D7 proteins the binding pocket is not as deep, and the ligand (magenta in Fig. 7a) extends out of the binding pocket and contacts residues in the C-terminal domain (not shown).
Figure 7.
Comparison of AeOBP22 with known structures. Cylinder representation of AeOBP22 in yellow (N and C-termini labeled in black) compared with (a) N-terminal OBP domain of An. stephensi D7 (blue) bound to Leukotriene C4 magenta (PDB 3NHI)67. For clarity, the C-terminal domain, which continues at the position labeled C+ in blue, is not shown. The linoleic acid bound to AeOBP22 is shown in cyan. (b) LmigOBP1 (grey) (PDB 4PT1)69. The difference in the positions of helices 5 and 7 are shown in red. (c) Phormia regina OBP56a bound to multiple molecules of palmitic acid (blue) (PDB 5DIC) (Ishida et al. not published). The multiple lysines and arginines that surround the pocket (grey) are shown in cyan. (d) An. gambiae OBP22 (bright blue) (PDB 3L4L) (Zhang and Ren, not published). The C-terminus of AgOBP22 is mostly unstructured (extends from position labeled with blue C+).
There are three structures of single domain OBPs that have an additional seventh C-terminal helix; An gambiae OBP7 (PDB 3R1O)68, Locusta migratoria OBP1 (LmigOBP1, PDB 4PT1)69 and the Mediterranean fruit fly, Ceratitis capitate OBP22 (PDB 6NHE)70. Of these, AeOBP22 is most similar to LmigOBP1 (Dali Z = 12.1) (Fig. 7b), however, helix 7 is significantly longer in LmigOPB1 and the angle between helices 6 and 7 differs by ~30° (red in Fig. 7b). Additionally, in AeOBP22 helices 4 and 5 are rotated out from the main body of the protein by ~28°. As a consequence, AeOBP22 has a longer, more extended pocket compared to that in LmigOBP1 (170 Å3).
AeOBP22 additionally shows structural conservation with the blowfly, Phormia regina, OBP56a (PregOBP56a, PDB 5DIC, Dali Z = 13.7) (Ishida et al. not published), which is proposed to transport fatty acids for feeding71. Helices 1 through 6 of PregOBP56a superimpose with AeOBP22 with an RMSD of 1.2 Å (Fig. 7c). However, PregOBP56a does not have a seventh helix. Instead, the N-terminal residues 1–5 occupy a similar position to the C-terminus of AeOBP22. The binding pocket of PregOBP56a is significantly larger (~530 Å3) and is lined by multiple lysine residues. This allows it to accommodate multiple ligands in a non-specific manner (Fig. 7c). This contrasts with AeOBP22, which has a hydrophobic pocket with Arg15 and Tyr46 positioned to make specific hydrogen bonds with carboxylic acid containing ligands (Fig. 3a).
AeOBP22 also has structural homology to An. gambiae OBP22 (AgamOBP22, PDB 3L4L, Dali Z = 12.5) (Zhang and Ren, not published) (Fig. 7d). AgamOBP22 differs in having a significantly shorter N-terminal region but simultaneously a much longer C-terminus, which is only partially observable in the deposited structures. This C-terminal region also forms a seventh α-helix, however the angle formed with helix-6 is more obtuse, ~116° compared to ~75°, seen in AeOBP22 and the binding pocket is relatively shallow and smaller (123 Å3) than that of AeOBP22.
Discussion
It is increasingly apparent that insect OBPs have diverse roles that extend beyond those of chemosensory signaling72. AeOBP22 is a prime example of this as it is expressed in multiple tissues3–7. Other OBPs expressed in multiple tissues include two related Helicoverpa spp., where it is proposed that HeOBP10 functions in both the delivery of an oviposition deterrent to fertilized eggs, and in the subsequent detection of that deterrent in the antenna23. Understanding the native ligands that interact with OBPs can provide insights into the underlying biology. Our results provide evidence that fatty acids are likely natural ligands for AeOBP22 and that the binding of long-chain fatty acids (C15-C20) is enhanced by a conformational change in the C-terminal tail that is critical to generate a high affinity binding site. Previously we showed that burial of a single methylene group from a ligand contributes ~1 Kcal mol−1 to the overall stability of the Drosophila OBP LUSH73 and so burial of a long alkyl chain can contribute significant binding energy leading to high affinity interactions.
Conformational changes associated with ligand binding have been demonstrated for multiple OBPs and PBPs37–39,61,63,64,74–78. These can stabilize the structure of the protein17,45,73,79, accommodate different ligands68,74,75,80,81 or displace ligands37,61,64,76–78,82,83. In Antheraea polyphemus PBP1, in the absence of ligand the C-terminal tail forms a long α-helix and occupies the binding site37. It functions to both displace ligands at lower pH but also contributes to high affinity ligand binding at higher pH84, even though it is displaced from the binding site, and so the mechanism for how it does this remains uncertain. The conformational change observed for AeOBP22 is more similar to the coil-to-helix transition observed for binding of norepinephrine to the C-terminal domain of Ae. aegypti D7 that results in capping of the ligand-binding pocket31. In the absence of ligand this C-terminal tail is highly dynamic. In contrast, the C-terminal tail of AeOBP22 is well ordered even in the absence of ligand (Fig. 3b). Therefore, we propose it functions as a selectivity filter for specific ligands of the appropriate chain length.
Previous studies of AeOBP224,59 showed that the length of the ligand was critical for binding, and also branching of the alkyl chain was detrimental to binding indicative of binding to a narrow pocket, consistent with our findings. However, in these studies the “best” ligands were compounds containing two aromatic rings. Such compounds would only partially occupy the ligand binding pocket we observe, and in agreement with this show binding affinities an order of magnitude weaker than long chain fatty acids. We cannot rule out the possibility that ligands other than fatty acids bind to AeOPB22, and indeed, our initial screens identified small molecules that induce conformational changes but only at much higher concentrations (>1 mM) and in a concentration dependent manner, suggesting that these if these compounds bind in vivo they likely require multiple ligands to induce a stable conformation of the protein. Our structural studies provide strong evidence that a negatively charged head group is required on the ligand given the number and arrangement of positively charged residues that surround the ligand binding pocket. Our preliminary screens failed to identify lyso-phosphatidic acid (LPA) (C16-LPA) as a ligand, as it did not produce the same stabilization of AeOBP22 we observed with free fatty acids. We now interpret this as because the overall chain length C16-LPA is too long (23 linear heavy atoms) to be accommodated in the binding pocket and indeed we found that fatty acids longer than C20 chains bind AeOBP22 with dramatically reduced affinities.
Fatty acids components of human sweat and skin and have critical roles in regulating mosquito behaviors. Dodecanoic acid and palmitoleic acid are strong oviposition attractants in Ae. aegypti, whilst the saturated C14–C16 and C18 acids are attractants at low concentrations but repellents at high concentrations85. In contrast, methyl ester derivatives were found to be oviposition deterrents85. Carboxylic acids have also been shown to synergize the effect of lactic acid as an attractant86. Carboxylic acids with C1–C3, C5–C8 and C13–C18 alkyl chains acids all increased attraction compared to lactic acid alone, whereas, C4 and C9–C12 had no effect or showed a decrease in attraction. Whilst other studies have suggested that C9 and C10 acids have a higher stimulatory effect compared to other fatty acids87. Our structural studies indicate that AeOBP22 retains the ability to bind to short chain carboxylic acids suggesting that AeOBP22 may have the ability to recognize different ligands in different tissues antennal AeOBP22 may modulate chemosensory responses to volatile short chain fatty acids, whether it functions as a transporter or a buffer of ligand concentration remains unknown20. Within the insect chemosensory system, fatty acids are endogenous components of the sensillar lymph fluid18, and these fatty acids can interact with both pheromones and PBPs on external sites to regulate pheromone accessibility and interactions between the PBP and the pheromone. In our NMR studies of AeOBP22 even at high fatty acid concentrations, we only observe a single binding site for the fatty acid and no evidence of other alternative sites of interaction.
In the proboscis AeOBP22 could be function to recognize fatty acids. However, its function in the salivary gland and male reproductive tissue is less clear. As a secreted protein in the salivary gland6,7 it is likely injected into the host during a blood meal. The closely related protein PregOBP56a from the blow-fly P. regina has been proposed to sequester and transport lipids for feeding71. It is unlikely that AeOBP22 plays such a role in the mosquito given the difference in the mechanisms of feeding. Rather, it seems that it must function to either transport a required ligand from the mosquito to the host or to sequester a signal present in the host. Similarly, given its expression in male reproductive tissue and its transmission to females, it would appear that it must be transporting or sequestering a pheromonal component. Our data suggest that in either case, any compound involved in these processes is likely to be a long chain negatively charged lipid structure, and the ability to undergo a conformational change that enhances binding may be an important evolutionary development that allows it to recognize low abundance compounds with high affinity.
Materials and Methods
Protein expression
The mature form of AeOBP22 lacking the N-terminal signal peptide was expressed and purified as previously described88,40.
NMR spectroscopy
NMR experiments were performed at 25 °C on a either a Varian 900 MHz DD2, Varian INOVA 600 MHz or Bruker Avance Neo 600 MHz spectrometer. Samples for NMR were dissolved in sodium phosphate (20 mM, pH 6.5) and 90% H2O/10% D2O, with DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) (80 μM) added as an internal reference 89,90. For ligand screening, protein concentration was 100 μM and ligands were added to a concentration of 200–500 μM. For chemical shift assignments, relaxation rate measurements and structure calculations, protein concentrations were in the 400–700 μM range. All NMR experiments were collected using non uniform sampling methods91 using the Poisson-gap sampling schemes implemented by Hyberts et al.92 with a sampling density of 25–50%. Data were processed using the istHMS package v211193,94 in combination with NmrPipe95 and analyzed using Ccpnmr Analysis v 2.4.296.
Chemical shift assignments
Chemical shift assignments for the complex with arachidonic acid and the backbone assignments for the apo-protein were previously reported40. Backbone 1H, 15N, 13Cα and 13Cβ assignments for nonadecanoic, linoleic acid (pH 4.5 and pH 6.5), and decanoic acids were made in the same way. All other backbone amide assignments were made by following chemical shift trajectories as a function of ligand concentration or chain length. Normalized chemical shift differences are reported as97,98
| 1 |
pH Titrations
Samples were dissolved in sodium citrate (20 mM, pH 6.5) and the pH adjusted using HCl and measured using a Lazar Ultra-M micro pH electrode (Lazar Labs, Los Angeles, CA).
NMR relaxation rates
15N R1, R1ρ and {1H}-15N heteronuclear NOEs for the apo-protein and the arachidonic acid complex were measured at 900 and 600 MHz with temperature compensation blocks, and the full set of relaxation delays were acquired prior to incrementation of the 15N evolution time99. Heteronuclear NOEs used a 5 second relaxation delay, with a saturation period of 3 seconds. For R1 measurements 11 relaxation delays were acquired spanning 0.01–1.2 seconds at 14 T and 0.01–1.8 s at 21 T. For R1ρ measurements, the spin lock field strength at 21 T was 2250 Hz and at 14 T it was 1829 Hz, and relaxation delays were arrayed over 30–210 ms. R1ρ values were converted into R2 values100 prior to analysis using the “d’Auvergne” protocol in the Relax software suite52,53,101–103.
NMR structure determination
NOE distance restraints were obtained from a simultaneous 15N/13C separated NOESY-HSQC104 with a 100 ms NOE mixing times. Intermolecular NOEs between the protein and ligand were recorded using a simultaneous 13C/15N F1-filtered, F3-edited NOESY-HSQC105,106 (150 ms mixing time), using samples recorded in both 90% H2O/10% D2O and a second that was exchanged into 99% D2O (pD 6.1). Intermolecular restraints were derived from those NOE correlations that were observed uniquely in the intermolecular NOE spectrum.
Backbone dihedral angle restraints were determined using Talos+107. Any dihedral angles that initially appeared over restrained (clustered at the extreme ends of the restraint) were removed from subsequent calculations. Unambiguous side-chain χ1 restraints were identified from 3JHAHB coupling constants measured as previously described108 in combination with NOE measurements. For structure calculations, the Karplus coefficients (A, B, and C)109 for 3JHAHB were parameterized as given in Perez et al.110.
Backbone amides involved in hydrogen-bonds were identified from an 1H-15N HSQC spectrum after the sample had been exchanged into 99% D2O based buffer. Acceptor groups for the H-bond restraints were identified after structure calculations had converged and these were included in the final iterations of refinement.
Structure calculations were performed using ARIA 2.3.2111,112 in combination with CNS-SOLVE (Version 1.2)113. NOE restraints lists were generated within CCPNMR analysis and divided into unambiguous and ambiguous restraint sets. Initial iterations used a soft-square well potential for the NOE restraints to identify violations in the restraint lists, and if violated these were subsequently treated as ambiguous restraints. The final rounds of calculations employed the log-harmonic potential in the final cooling stage114 in combination with a modified “soft” force field and updated weighting parameters115. In the final iteration, a total of 100 structures were determined, and the top 50 were refined in water and the top 30 of these selected for analysis based on their total energy as described114. Superposition of structures and calculation of RMSD were performed using the suppose program from Ambertools116.
X-ray crystallography
Crystallization
For crystallization trials, AeOBP22 was exchanged into 4-(2-hydroxyethyl)-1-piperazine-ethane-sulfonic acid (HEPES) buffer at pH 7.5. The protein was concentrated to 9–12 mg ml-1 and incubated with ligands at room temperature overnight prior to crystallization trials. Crystals were grown by sitting drop vapor-diffusion.
Initial crystals were obtained in the P3121 space group in the presence of benzaldehyde (2 mM), by mixing the protein solution in a 1:1 ratio with a precipitant solution containing sodium-potassium tartrate (0.2 M), sodium citrate (0.1 M) pH 5.0 and ammonium sulfate (2 M). A tantalum bromide derivative was obtained by soaking crystals with a 2 mM Ta6Br12 solution (Jena Biosciences) for 2–3 days. Crystals of the complexes in the P31 space group were obtained at 18 °C through optimization of an initial hit containing ammonium sulfate (1.2–1.4 M), citric acid (0.1 M), and varying the pH over 5.5–5.7, and optimized by addition of sodium chloride to the well solution across a concentration range of 1.0–1.5 M. Crystals in the C121 space group were obtained using PEG3550 (12% w/v) and HEPES (0.1 M) at pH 7.5. For the C18:2 complex the precipitant solution additionally contained cobalt chloride (4 mM), cadmium chloride (4 mM) and magnesium chloride (4 mM). Arachidonic acid complex crystals were obtained with cobalt chloride (6 mM), cadmium chloride (10 mM) and magnesium chloride (6 mM). The apo-protein crystal was obtained with cobalt chloride (6 mM), cadmium chloride (6 mM) and sodium chloride (6 mM).
Data collection
X-ray diffraction data was collected at the Molecular Biology Consortium, Beamline 4.2.2. at the Advanced Light Source, LBNL, Berkley CA (crystal form 1 and 2) and the University of Colorado School of Medicine Biomolecular X-ray Crystallography Center (crystal form 3) using a Rigaku MicroMax™ 007 HF generator with a copper anode and equipped with a Rigaku PILATUS3 R 200 K detector.
Structure solution and refinement
The initial structure of AeOBP22 was solved using SAD methods of a Ta6Br12 soaked crystals in the P3121 space group collected at a wavelength of 1.254 Å. Data were integrated and scaled using XDS117,118 and refined to a resolution of 2.59 Å using Phenix119 to locate heavy atoms sites and generate initial maps. A native data set collected at 1.0 Å on the same crystal was solved using Phenix119 with model rebuilding in Coot120 and refined to a resolution of 1.9 Å.
Crystal structures of the complexes in the P31 space group were solved by molecular replacement121 using the structure obtain from the Ta6Br12 crystal as an initial search model. Data were collected at both the ALS beamline 4.2.2 and on the home-source, which was processed with HKL3000122. These crystals are pathologically twinned (~46%), and data were refined using intensity based twin refinement in Refmac123 within CCP4124,125. Structures of the complexes in the C121 space group were solved by cobalt or cadmium SAD methods using data sets collected in house at a wavelength of 1.5418 Å, structures were solved using Phenix and subsequently refined directly against the SAD data in Refmac123.
Ligand binding measurements
Affinity of AeOBP22 for 5-(N-dodecanoyl)-aminofluorescein (DAF)
All fluorescence experiments were recorded at 25 °C on a Horiba Fluorolog 3–1–1 spectrofluorometer. DAF spectra were acquired using an excitation wavelength of 490 nm and the emission recorded between 500 and 560 nm at 1 nm intervals with an integration time of 1 s and slit widths of 2 nm. Stock solutions of DAF (Thermo Fisher, Batch D-109) were prepared in methanol with potassium hydroxide (0.2 M) and the concentration measured by UV spectroscopy using an extinction coefficient of 88000 M−1 cm−1 at 497 nm provided in the certificate of analysis. A 1:100 dilution was made into HEPES (50 mM, pH 8.1) and the sample vortexed and checked by UV to verify the DAF concentration. A solution of DAF (~80–100 nM) was titrated with increasing AeOBP22 to a final concentration of ~4 μM. The fluorescence intensity (cps) at the emission maxima (513 nm) was plotted against the total concentration of AeOBP22 and the KD for DAF was determined by fitting the raw fluorescence intensity for each replicate to Eq. (1):
| 2 |
where Fi is the measured fluorescence, F0 is the initial fluorescence in the absence of any protein, PT is the total protein, LT is the total concentration of DAF, and Fmin is the residual fluorescence after maximal quenching by the protein.
Binding affinity for fatty acids
Binding affinities were determined by monitoring the recovery of DAF florescence as a function of fatty acid concentration. Stock solutions of fatty acid were made by dissolving the fatty acid in 100% DMSO, and then making a 1:1000 dilution into HEPES (50 mM pH 8.1). Samples were vortexed prior to each titration point. The protein/DAF solution contained fixed concentrations of DAF (100 nM) and AeOBP22 (1000 nM). Background controls involved titration of the same DAF solution with solvent or fatty acid which showed no significant effect on the fluorescence intensity of DAF.
As there is a single binding site for the fatty acid and the effect of fluorescence quenching can be completely reversed by increasing concentrations of fatty acids, we concluded that the fatty acid is directly competitive with binding of the DAF reporter. Therefore, the term for KD in Eq. 2 can be replaced with Eq. 3.
| 3 |
where I is the concentration of the competitor and KI is the dissociation constant of the competitor. The emission maxima were fit using Eq. 2 substituted into Eq. 1, with the concentration of the protein and DAF fixed at their known concentrations.
Supplementary Information
Acknowledgements
This work was supported by NIH/NIAID grant RO1 AI121253 to DNMJ. The operation of the Structural Biology facilities at CU School of Medicine is supported in part by the University of Colorado Cancer Center Support Grant (NIH 5P30-CA046934). We thank Dr George Dimopoulos for AeOBP22 cDNA and discussions.
Author contributions
D.N.M.J. conceived the experiments; J.W. and E.J.M. performed experiments; J.W., E.J.M., J.C.N. and D.N.M.J. all collected and analyzed X-ray data; J.W., E.J.M. and D.N.M.J. collected and analyzed the N.M.R. data; and J.W. and D.N.M.J. wrote the manuscript.
Data availability
Coordinates for the AeOBP22 and its complexes have been deposited in the Protein Data Bank (https://www.rcsb.org/) with the accession numbers: 6OTL – TaBr Complex (SAD); 6P2E – TaBr (Native); 6OMW – palmitoleic acid; 6OPB – eicosanoic acid; 6OG0 – apo state; 6OGH – linoleic acid; 6OII – arachidonic acid. 6NBN – NMR structure with arachidonic acid. NMR chemical shift assignments and structural restraints have been deposited with the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/) with accession numbers 27724 and 30550.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60242-9.
References
- 1.Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 3.Sim S, Ramirez JL, Dimopoulos G. Dengue Virus Infection of the Aedes aegypti Salivary Gland and Chemosensory Apparatus Induces Genes that Modulate Infection and Blood-Feeding Behavior. PLoS Path. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S, et al. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem. Biophys. Res. Commun. 2008;372:464–468. doi: 10.1016/j.bbrc.2008.05.064. [DOI] [PubMed] [Google Scholar]
- 5.Sirot LK, et al. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect Biochem. Mol. Biol. 2008;38:176–189. doi: 10.1016/j.ibmb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhawan R, et al. Mosquito-Borne Diseases and Omics: Salivary Gland Proteome of the Female Aedes aegyptiMosquito. OMICS: J. Integr. Biol. 2017;21:45–54. doi: 10.1089/omi.2016.0160. [DOI] [PubMed] [Google Scholar]
- 7.Dhawan R, et al. Data from salivary gland proteome analysis of female Aedes aegypti Linn. Data Brief. 2017;13:274–277. doi: 10.1016/j.dib.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nat. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 9.Kim M-S, Repp A, Smith DP. LUSH Odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genet. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is Required for Activity of Pheromone-Sensitive Neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Krieger J, Breer H. Olfactory reception in invertebrates. Sci. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- 12.Sun JS, Xiao S, Carlson JR. The diverse small proteins called odorant-binding proteins. Open. Biol. 2018;8:180208. doi: 10.1098/rsob.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forstner M, Breer H, Krieger J. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci. 2009;5:745–757. doi: 10.7150/ijbs.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 2007;25:2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- 15.Grosse-Wilde E, Svatos A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses. 2006;31:547–555. doi: 10.1093/chemse/bjj059. [DOI] [PubMed] [Google Scholar]
- 16.Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc. Natl Acad. Sci. USA. 2006;103:16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nardella J, Terrado M, Honson NS, Plettner E. Endogenous fatty acids in olfactory hairs influence pheromone binding protein structure and function in Lymantria dispar. Arch. Biochem. Biophys. 2015;579:73–84. doi: 10.1016/j.abb.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Honson N, Johnson MA, Oliver JE, Prestwich GD, Plettner E. Structure-Activity Studies with Pheromone-binding Proteins of the Gypsy Moth, Lymantria dispar. Chem. Senses. 2003;28:479–489. doi: 10.1093/chemse/28.6.479. [DOI] [PubMed] [Google Scholar]
- 20.Larter, N. K., Sun, J. S. & Carlson, J. R. Organization and function of Drosophila odorant binding proteins. eLife510.7554/eLife.20242. (2016). [DOI] [PMC free article] [PubMed]
- 21.Rothemund S, Liou YC, Davies PL, Krause E, Sonnichsen FD. A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands. Structure. 1999;7:1325–1332. doi: 10.1016/S0969-2126(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 22.Takemori N, Yamamoto MT. Proteome mapping of the Drosophila melanogaster male reproductive system. Proteom. 2009;9:2484–2493. doi: 10.1002/pmic.200800795. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y-L, Huang L-Q, Pelosi P, Wang C-Z. Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS One. 2012;7:e30040. doi: 10.1371/journal.pone.0030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scolari F, et al. The Spermatophore in Glossina morsitans morsitans: Insights into Male Contributions to Reproduction. Sci. Rep. 2016;6:20334. doi: 10.1038/srep20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amenya DA, et al. Proteomics reveals novel components of the Anopheles gambiae eggshell. J. Insect Physiol. 2010;56:1414–1419. doi: 10.1016/j.jinsphys.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa-da-Silva AL, Kojin BB, Marinotti O, James AA, Capurro ML. Expression and accumulation of the two-domain odorant-binding protein AaegOBP45 in the ovaries of blood-fed Aedes aegypti. Parasit. Vectors. 2013;6:364. doi: 10.1186/1756-3305-6-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marinotti O, et al. Integrated proteomic and transcriptomic analysis of the Aedes aegypti eggshell. BMC Dev. Biol. 2014;14:15. doi: 10.1186/1471-213X-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji R, et al. Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence. PLoS One. 2013;8:e79612. doi: 10.1371/journal.pone.0079612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celorio-Mancera Mde L, et al. Chemosensory proteins, major salivary factors in caterpillar mandibular glands. Insect Biochem. Mol. Biol. 2012;42:796–805. doi: 10.1016/j.ibmb.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- 31.Calvo E, Mans BJ, Ribeiro JMC, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc. Natl Acad. Sci. USA. 2009;106:3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mans BJ, Calvo E, Ribeiro JM, Andersen JF. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J. Biol. Chem. 2007;282:36626–36633. doi: 10.1074/jbc.M706410200. [DOI] [PubMed] [Google Scholar]
- 33.Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J. Biol. Chem. 2002;277:27651–27658. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro JM. Blood-feeding in mosquitoes: probing time and salivary gland anti-haemostatic activities in representatives of three genera (Aedes, Anopheles, Culex) Med. Vet. Entomol. 2000;14:142–148. doi: 10.1046/j.1365-2915.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- 35.Conway MJ, et al. Aedes aegypti D7 Saliva Protein Inhibits Dengue Virus Infection. PLoS Neglected Tropical Dis. 2016;10:e0004941. doi: 10.1371/journal.pntd.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conway MJ, et al. Mosquito saliva serine protease enhances dissemination of dengue virus into the mammalian host. J. Virol. 2014;88:164–175. doi: 10.1128/JVI.02235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katre UV, Mazumder S, Prusti RK, Mohanty S. Ligand binding turns moth pheromone-binding protein into a pH sensor: effect on the Antheraea polyphemus PBP1 conformation. J. Biol. Chem. 2009;284:32167–32177. doi: 10.1074/jbc.M109.013383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zubkov S, Gronenborn AM, Byeon IJ, Mohanty S. Structural consequences of the pH-induced conformational switch in A.polyphemus pheromone-binding protein: mechanisms of ligand release. J. Mol. Biol. 2005;354:1081–1090. doi: 10.1016/j.jmb.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 39.Mohanty S, Zubkov S, Gronenborn AM. The solution NMR structure of Antheraea polyphemus PBP provides new insight into pheromone recognition by pheromone-binding proteins. J. Mol. Biol. 2004;337:443–451. doi: 10.1016/j.jmb.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Jones DNM, Wang J, Murphy EJ. Complete NMR chemical shift assignments of odorant binding protein 22 from the yellow fever mosquito, Aedes aegypti, bound to arachidonic acid. Biomol. NMR Assign. 2019;13:187–193. doi: 10.1007/s12104-019-09875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plettner E, Lazar J, Prestwich EG, Prestwich GD. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar. Biochem. 2000;39:8953–8962. doi: 10.1021/bi000461x. [DOI] [PubMed] [Google Scholar]
- 43.Briand L, Nespoulous C, Huet JC, Takahashi M, Pernollet JC. Ligand binding and physico-chemical properties of ASP2, a recombinant odorant-binding protein from honeybee (Apis mellifera L.) Eur. J. Biochem. 2001;268:752–760. doi: 10.1046/j.1432-1327.2001.01927.x. [DOI] [PubMed] [Google Scholar]
- 44.Wogulis M, Morgan T, Ishida Y, Leal WS, Wilson DK. The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism. Biochem. Biophys. Res. Commun. 2006;339:157–164. doi: 10.1016/j.bbrc.2005.10.191. [DOI] [PubMed] [Google Scholar]
- 45.Davrazou F, Dong E, Murphy EJ, Johnson HT, Jones DN. New insights into the mechanism of odorant detection by the malaria-transmitting mosquito Anopheles gambiae. J. Biol. Chem. 2011;286:34175–34183. doi: 10.1074/jbc.M111.274712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsitsanou KE, et al. Crystal and solution studies of the “Plus-C” odorant-binding protein 48 from Anopheles gambiae: control of binding specificity through three-dimensional domain swapping. J. Biol. Chem. 2013;288:33427–33438. doi: 10.1074/jbc.M113.505289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao H, et al. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell. Mol. Life Sci. 2011;68:1799–1813. doi: 10.1007/s00018-010-0539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochem. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 49.Kroenke CD, Loria JP, Lee LK, Rance M, Palmer AG. Longitudinal and transverse H-1-N-15 dipolar N-15 chemical shift anisotropy relaxation interference: Unambiguous determination of rotational diffusion tensors and chemical exchange effects in biological macromolecules. J. Am. Chem. Soc. 1998;120:7905–7915. doi: 10.1021/ja980832l. [DOI] [Google Scholar]
- 50.Luginbuhl P, Wuthrich K. Semi-classical nuclear spin relaxation theory revisited for use with biological macromolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2002;40:199–247. doi: 10.1016/S0079-6565(01)00043-7. [DOI] [Google Scholar]
- 51.Cavanagh, J., Fairbrother, W. J., Palmer III, A. G., Rance, M. & Skelton, N. Protein NMR Spectroscopy: Principles and Practice. 2nd Vol. (ed. (Elsevier, 2007).
- 52.d’Auvergne EJ, Gooley PR. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR. 2008;40:107–119. doi: 10.1007/s10858-007-9214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.d’Auvergne EJ, Gooley PR. Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J. Biomol. NMR. 2008;40:121–133. doi: 10.1007/s10858-007-9213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bieri M, d’Auvergne EJ, Gooley PR. relaxGUI: a new software for fast and simple NMR relaxation data analysis and calculation of ps-ns and mus motion of proteins. J. Biomol. NMR. 2011;50:147–155. doi: 10.1007/s10858-011-9509-1. [DOI] [PubMed] [Google Scholar]
- 55.Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim IH, et al. A mosquito hemolymph odorant-binding protein family member specifically binds juvenile hormone. J. Biol. Chem. 2017;292:15329–15339. doi: 10.1074/jbc.M117.802009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou JJ, et al. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: evidence for odour recognition and discrimination. FEBS Lett. 2004;558:23–26. doi: 10.1016/S0014-5793(03)01521-7. [DOI] [PubMed] [Google Scholar]
- 58.Murphy EJ, Booth JC, Davrazou F, Port AM, Jones DN. Interactions of Anopheles gambiae odorant-binding proteins with a human-derived repellent: implications for the mode of action of n,n-diethyl-3-methylbenzamide (DEET) J. Biol. Chem. 2013;288:4475–4485. doi: 10.1074/jbc.M112.436386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang G, Winberg G, Ren H, Zhang S. Expression, purification and functional analysis of an odorant binding protein AaegOBP22 from Aedes aegypti. Protein Expr. Purif. 2011;75:165–171. doi: 10.1016/j.pep.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 60.Murphy, E. J., Booth, J. C., Davrazou, F., Port, A. M. & Jones, D. N. Interactions of Anopheles gambiae odorant binding proteins with a human-derived repellent: implications for the mode of action of DEET. J Biol Chem (2012). [DOI] [PMC free article] [PubMed]
- 61.Wojtasek H, Leal W. Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. J. Biol. Chem. 1999;274:30950–30956. doi: 10.1074/jbc.274.43.30950. [DOI] [PubMed] [Google Scholar]
- 62.Damberger F, et al. NMR characterization of a pH-dependent equilibrium between two folded solution conformations of the pheromone-binding protein from Bombyx mori. Protein Sci. 2000;9:1038–1041. doi: 10.1110/ps.9.5.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 2000;7:143–151. doi: 10.1016/S1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- 64.Lautenschlager C, Leal WS, Clardy J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem. Biophys. Res. Commun. 2005;335:1044–1050. doi: 10.1016/j.bbrc.2005.07.176. [DOI] [PubMed] [Google Scholar]
- 65.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holm, L., Kaariainen, S., Wilton, C. & Plewczynski, D. Using Dali for structural comparison of proteins. Curr Protoc BioinformaticsChapter 5, Unit 5 5, (2006). [DOI] [PubMed]
- 67.Alvarenga PH, et al. The function and three-dimensional structure of a thromboxane A2/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite. PLoS Biol. 2010;8:e1000547. doi: 10.1371/journal.pbio.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lagarde A, et al. The crystal structure of odorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. J. Mol. Biol. 2011;414:401–412. doi: 10.1016/j.jmb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Zheng J, et al. Crystal structure of the Locusta migratoria odorant binding protein. Biochem. Biophys. Res. Commun. 2015;456:737–742. doi: 10.1016/j.bbrc.2014.12.048. [DOI] [PubMed] [Google Scholar]
- 70.Falchetto, M. et al. Structural and biochemical evaluation of Ceratitis capitata OBP22 affinity for odorants involved in inter-sex communication. Insect Mol. Biol., imb.1255, (2018). [DOI] [PubMed]
- 71.Ishida Y, Ishibashi J, Leal WS. Fatty Acid Solubilizer from the Oral Disk of the Blowfly. PLoS One. 2013;8:e51779. doi: 10.1371/journal.pone.0051779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelosi P, Iovinella I, Zhu J, Wang G, Dani FR. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2017;6:1669. doi: 10.1111/brv.12339. [DOI] [PubMed] [Google Scholar]
- 73.Bucci BK, Kruse SW, Thode AB, Alvarado SM, Jones DNM. Effect of n-alcohols on the structure and stability of the Drosophila odorant binding protein LUSH. Biochem. 2006;45:1693–1701. doi: 10.1021/bi0516576. [DOI] [PubMed] [Google Scholar]
- 74.Pesenti ME, et al. Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release. J. Mol. Biol. 2009;390:981–990. doi: 10.1016/j.jmb.2009.05.067. [DOI] [PubMed] [Google Scholar]
- 75.Pesenti ME, et al. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J. Mol. Biol. 2008;380:158–169. doi: 10.1016/j.jmb.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 76.Xu X, et al. NMR structure of navel orangeworm moth pheromone-binding protein (AtraPBP1): implications for pH-sensitive pheromone detection. Biochem. 2010;49:1469–1476. doi: 10.1021/bi9020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu W, Xu X, Leal WS, Ames JB. Extrusion of the C-terminal helix in navel orangeworm moth pheromone-binding protein (AtraPBP1) controls pheromone binding. Biochem. Biophys. Res. Commun. 2011;404:335–338. doi: 10.1016/j.bbrc.2010.11.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.di Luccio E, Ishida Y, Leal WS, Wilson DK. Crystallographic observation of pH-induced conformational changes in the Amyelois transitella pheromone-binding protein AtraPBP1. PLoS One. 2013;8:e53840. doi: 10.1371/journal.pone.0053840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kruse SW, Zhao R, Smith DP, Jones DN. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat. Struct. Biol. 2003;10:694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lescop E, Briand L, Pernollet J-C, Guittet E. Structural basis of the broad specificity of a general odorant-binding protein from honeybee. Biochem. 2009;48:2431–2441. doi: 10.1021/bi802300k. [DOI] [PubMed] [Google Scholar]
- 81.Spinelli S, et al. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem. Mol. Biol. 2012;42:41–50. doi: 10.1016/j.ibmb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 82.Damberger FF, Ishida Y, Leal WS, Wüthrich K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J. Mol. Biol. 2007;373:811–819. doi: 10.1016/j.jmb.2007.07.078. [DOI] [PubMed] [Google Scholar]
- 83.Horst R, et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc. Natl. Acad. Sci. USA. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katre UV, Mazumder S, Mohanty S. Structural Insights into the Ligand Binding and Releasing Mechanism of Antheraea polyphemusPheromone-Binding Protein 1: Role of the C-Terminal Tail. Biochem. 2013;52:1037–1044. doi: 10.1021/bi301393v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ganesan K, Mendki MJ, Suryanarayana MVS, Prakash S, Malhotra RC. Studies of Aedes aegypti (Diptera: Culicidae) ovipositional responses to newly identified semiochemicals from conspecific eggs. Aust. J. Entomol. 2006;45:75–80. doi: 10.1111/j.1440-6055.2006.00513.x. [DOI] [Google Scholar]
- 86.Bosch OJ, Geier M, Boeckh J. Contribution of fatty acids to olfactory host finding of female Aedes aegypti. Chem. Senses. 2000;25:323–330. doi: 10.1093/oxfordjournals.chemse.a014042. [DOI] [PubMed] [Google Scholar]
- 87.Ponnusamy L, et al. Identification of bacteria and bacteria-associated chemical cues that mediate oviposition site preferences by Aedes aegypti. Proc. Natl Acad. Sci. USA. 2008;105:9262–9267. doi: 10.1073/pnas.0802505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prestwich GD. Bacterial expression and photoaffinity labeling of a pheromone binding protein. Prot. Sci. 1993;2:420–428. doi: 10.1002/pro.5560020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wishart DS, et al. H-1, C-13 and N-15 Chemical-Shift Referencing in Biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 90.Markley JL, et al. Recommendations for the presentation of NMR structures of proteins and nucleic acids - (IUPAC Recommendations 1998) Pure Appl. Chem. 1998;70:117–142. doi: 10.1351/pac199870010117. [DOI] [Google Scholar]
- 91.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. Exponential sampling, an alternative method for sampling in two-dimensional NMR experiments. J. Magn. Reson. 1987;73:69–77. [Google Scholar]
- 92.Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hyberts SG, Arthanari H, Robson SA, Wagner G. Perspectives in magnetic resonance: NMR in the post-FFT era. J. Magn. Reson. 2014;241:60–73. doi: 10.1016/j.jmr.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 96.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 97.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 98.Schumann FH, et al. Combined chemical shift changes and amino acid specific chemical shift mapping of protein–protein interactions. J. Biomol. NMR. 2007;39:275–289. doi: 10.1007/s10858-007-9197-z. [DOI] [PubMed] [Google Scholar]
- 99.Dayie KT, Wagner G. Relaxation-Rate Measurements for N-15-H-1 Groups with Pulsed-Field Gradients and Preservation of Coherence Pathways. J. Magn. Reson., Ser. A. 1994;111:121–126. doi: 10.1006/jmra.1994.1236. [DOI] [Google Scholar]
- 100.Palmer AG, Massi F. Characterization of the Dynamics of Biomacromolecules Using Rotating-Frame Spin Relaxation NMR Spectroscopy. Chem. Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 101.d’Auvergne EJ, Gooley PR. Set theory formulation of the model-free problem and the diffusion seeded model-free paradigm. Mol. Biosyst. 2007;3:483–494. doi: 10.1039/b702202f. [DOI] [PubMed] [Google Scholar]
- 102.d’Auvergne EJ, Gooley PR. Model-free model elimination: a new step in the model-free dynamic analysis of NMR relaxation data. J. Biomol. NMR. 2006;35:117–135. doi: 10.1007/s10858-006-9007-z. [DOI] [PubMed] [Google Scholar]
- 103.d’Auvergne EJ, Gooley PR. The use of model selection in the model-free analysis of protein dynamics. J. Biomol. NMR. 2003;25:25–39. doi: 10.1023/A:1021902006114. [DOI] [PubMed] [Google Scholar]
- 104.Vögeli B, Güntert P, Riek R. Multiple-state ensemble structure determination from eNOE spectroscopy. Mol. Phys. 2013;111:437–454. doi: 10.1080/00268976.2012.728257. [DOI] [Google Scholar]
- 105.Pascal SM, Muhandiram DR, Yamazaki T, Formankay JD, Kay LE. Simultaneous Acquisition of N-15-Edited and C-13-Edited Noe Spectra of Proteins Dissolved in H2o. J. Magn. Reson., Ser. B. 1994;103:197–201. doi: 10.1006/jmrb.1994.1031. [DOI] [Google Scholar]
- 106.Zwahlen C, et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage lambda N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997;119:6711–6721. doi: 10.1021/ja970224q. [DOI] [Google Scholar]
- 107.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Born Alexandra, Henen Morkos, Nichols Parker, Wang Jing, Jones David, Vögeli Beat. Efficient Stereospecific Hβ2/3 NMR Assignment Strategy for Mid-Size Proteins. Magnetochemistry. 2018;4(2):25. doi: 10.3390/magnetochemistry4020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karplus M. Vicinal Proton Coupling in Nuclear Magnetic Resonance. J. Am. Chem. Soc. 1963;85:2870–2871. doi: 10.1021/ja00901a059. [DOI] [Google Scholar]
- 110.Perez C, Lohr F, Ruterjans H, Schmidt JM. Self-consistent Karplus parametrization of 3J couplings depending on the polypeptide side-chain torsion chi1. J. Am. Chem. Soc. 2001;123:7081–7093. doi: 10.1021/ja003724j. [DOI] [PubMed] [Google Scholar]
- 111.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinforma. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 112.Linge JP, O’Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/S0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 113.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;D54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- 114.Nilges M, et al. Accurate NMR Structures Through Minimization of an Extended Hybrid Energy. Structure. 2008;16:1305–1312. doi: 10.1016/j.str.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 115.Mareuil, F., Malliavin, T. E., Nilges, M. & Bardiaux, B. Improved reliability, accuracy and quality in automated NMR structure calculation with ARIA. J. Biomol. NMR, 1–14 (2015). [DOI] [PMC free article] [PubMed]
- 116.AMBER 2018 (University of California, San Francisco, 2018).
- 117.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 121.Brünger AT. Extension of molecular replacement: A new strategy based on Patterson correlation refinement. Acta Crystallogr. 1990;A46:472–475. [Google Scholar]
- 122.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 123.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 124.Bailey S. The CCP4 Suite - Programs for Protein Crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444993011898. [DOI] [PubMed] [Google Scholar]
- 125.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates for the AeOBP22 and its complexes have been deposited in the Protein Data Bank (https://www.rcsb.org/) with the accession numbers: 6OTL – TaBr Complex (SAD); 6P2E – TaBr (Native); 6OMW – palmitoleic acid; 6OPB – eicosanoic acid; 6OG0 – apo state; 6OGH – linoleic acid; 6OII – arachidonic acid. 6NBN – NMR structure with arachidonic acid. NMR chemical shift assignments and structural restraints have been deposited with the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/) with accession numbers 27724 and 30550.