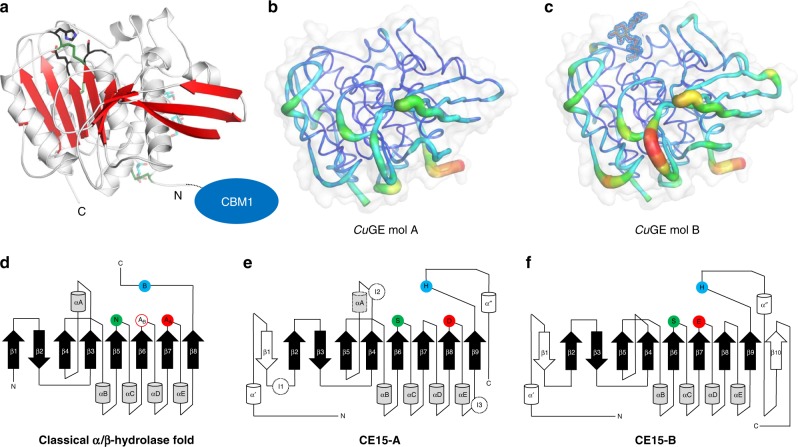
Fig. 3. Overall CuGE structure, CE15 subgroups, and relation to the classical α/β-hydrolase fold.
a Structure of the catalytic domain of CuGE (residues 79–458) with selected residues shown as sticks: the catalytic triad (S270, E293, and H404, black), disulfide bridges (C81–C116, C269–C405, C391–C377, green), and carbohydrates (cyan), indicating the location of N- and O-glycosylation sites (N104-NAG, S86-MAN, and T87-MAN). The break in strand β3 is caused by a proline residue (P169) in CuGE. b, c The two crystallographically independent molecules A and B from the dΔS270A:Um4X complex colored by temperature factors to visualize flexibility. b Molecule A. c Ligand-bound molecule B. d Topology diagram for a classical α/β-hydrolase fold. The illustration is based on information in Ollis et al.23, Carr et al.24, and Dimitrou et al.25. The location of the catalytic residues is shown (nucleophile serine in green, base histidine in blue, and acid glutamate in red). The α/β-hydrolase group A and B are distinguished by different configurations of the catalytic triad with the catalytic acid located either in the turn after β7 (AA) or after β6 (AB), respectively25. e Topology diagram for CE15-A. I1-I3 indicate inserts observed in bacterial CE15-A structures13–15. f Topology diagram for CE15-B. For clarity, only the major secondary structure elements have been included in the topology diagrams in e, f; structures from both CE15 subgroups comprise additional small secondary structure elements, including multiple helical elements inserted after the strands β5 and β7 (Supplementary Fig. 4).