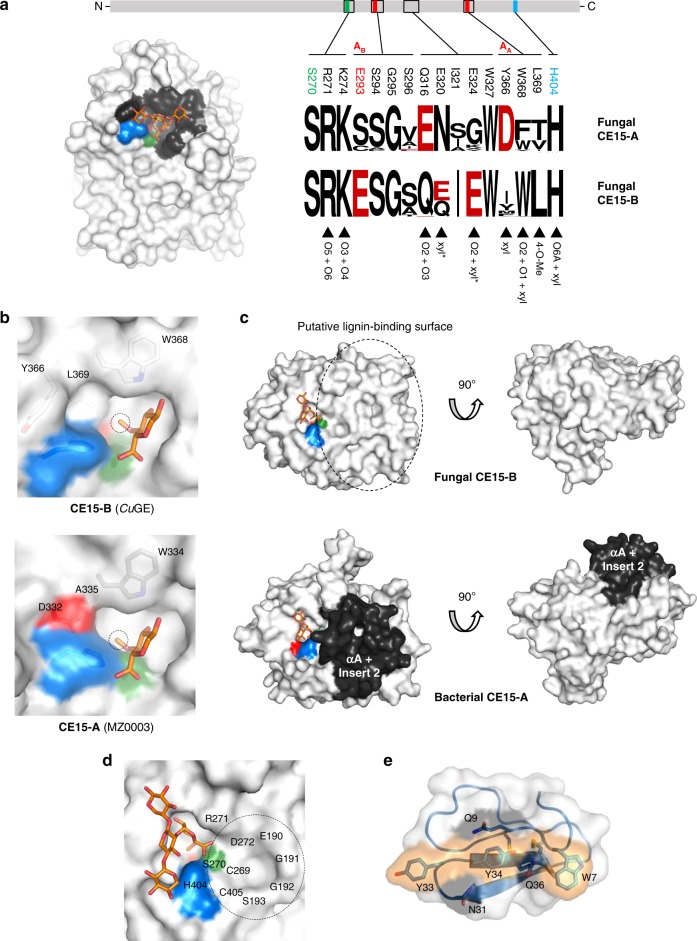
Fig. 6. Conservation of structural features implicated in recognition of natural substrates by fungal CE15 enzymes.
a Conservation of the carbohydrate-binding site. Residues within 5 Å from the XUm4X fragment are mapped onto the surface of CuGE. Corresponding sequence logos50 are shown for the fungal subgroups CE15-A and CE15-B, respectively. The role(s) played by the individual residues in the CuGE complexes with aldouronic acids are summarized. b Distinct environments around the 4-O-Methyl group caused by the different configurations of the catalytic triads in CE15-A and CE15-B. The ligand from the dΔS270A:XUm4XX-OH structure is superimposed on the MZ0003 structure (6EHN)13. For clarity, only the 4-O-methyl-glucuronoyl-part of the ligand is shown. c Surface representations of CuGE (CE15-B) and MZ0003 (CE15-A), illustrating differences in surface topology. Notably, the αA-helix and “insert 2” are only present in the bacterial members of CE15-A. In all fungal enzymes (both CE15-A and CE15-B), a deletion in this region results in a very open, flat, and accessible-binding site. d Putative monolignol binding site. e CuGE-CBM1 homology model showing the conserved residues implicated in the adsorption onto insoluble substrates.