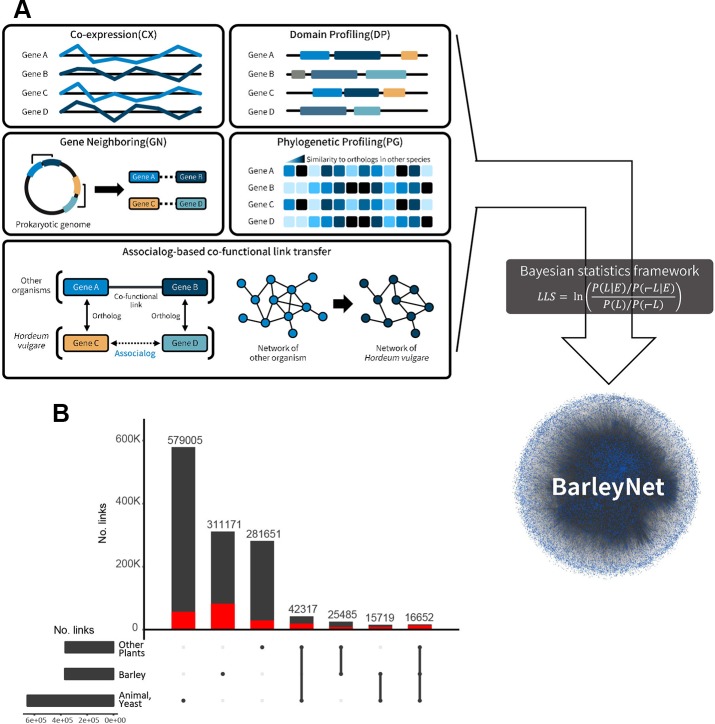
Figure 1.
Overview of BarleyNet. (A) BarleyNet was constructed by integrating functional associations between barley genes inferred from the co-expression of genes (CX), gene neighborhood (GN), association of protein domain composition profiles (DP), phylogenetic profile association (PG), and those transferred from 21 networks previously constructed for other species based on functional association between orthologous proteins (associalog). (B) Summary of BarleyNet edge information with UpSet visualization. Network edges were classified into three groups based on the species of origin of the inferred co-functional association: “barley,” “other plants” and “animals or yeast.” The bar height represents the number of BarleyNet links for each species group or their combination. The red bar represents the number of links that are 20 fold more likely than gene pairs by random (i.e., high confidence links).