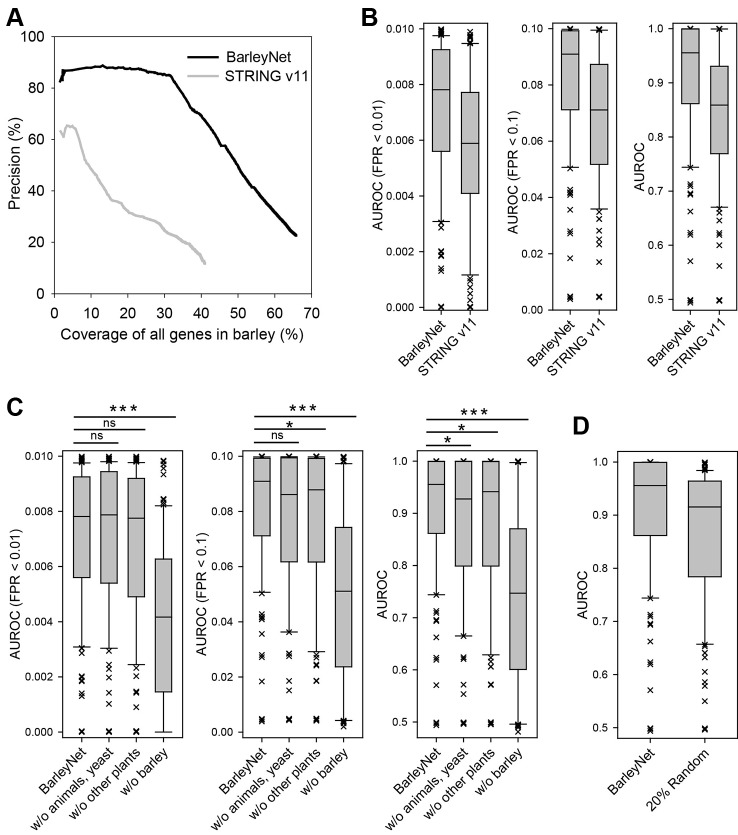
Figure 2.
Assessment of BarleyNet and a network of barley genes by STRING database. (A) The quality of the networks was evaluated based on precision for gene pairs that have the same GOBP terms by agriGO annotations and coverage of all barley genes. BarleyNet showed substantially higher precision than the network of barley genes by the STRING v11 database considering the entire range of coverage. (B) Comparison of area under receiver operating characteristic curve (AUROC) of 122 pathway gene sets derived from Plant Reactome database. Box-and-whisker plots represent 10%, 25%, median, 75%, and 90% of 122 AUROC scores. The same AUROC analyses were conducted until 1%, 10%, and 100% of false positive rate (FPR) were reached. BarleyNet showed a significantly higher prediction power than the STRING database barley gene network in all FPR ranges (P < 0.001, Wilcoxon signed rank test). (C) AUROC analyses were conducted as for (B) with BarleyNet and the following “dropout” networks by excluding links from animals and yeast (w/o animals, yeast), by excluding links from Arabidopsis, rice, and maize (w/o other plants), and by excluding links from barley (w/o barley). ns, not significant; *, P < 0.05; ***, P < 0.001 by Wilcoxon signed rank test. (D) AUROC analyses were conducted as for (B) with 100 networks in which 20% of BarleyNet links were randomized. Average AUROC scores for the 122 pathways gene sets across 100 networks are represented in the Box-and-whisker plot for randomized networks.