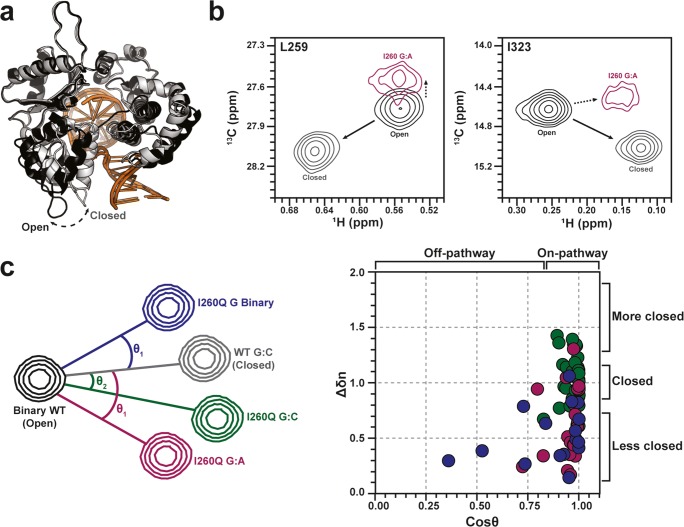
Fig. 6.
a X-ray crystal structures of open (black) and closed (gray) pol β. DNA molecules are shown in orange shades. b NMR chemical shift signatures of the open (black, WT without nucleotide), closed (gray, WT with matched nucleotide), and “off-pathway” (purple, I260Q mutant with mismatched nucleotide) structural states of pol β. c Schematic representation of chemical shift vector analysis reported by Liptak et al., accounting for various degrees of enzymatic closure around matched and mismatched nucleotides in WT and I260Q mutant pol β. WT pol β adopts its native, closed structure upon matched dNTP binding (black, gray), while the cancer-associated I260Q variant populates altered structures with various dNTPs (red, blue, green) that deviate from the WT pathway. The normalized vector magnitude was calculated as ∆δn = ∆δexptl/∆δref, where ∆δref is the WT trajectory. The degree of I260Q pol β closure and its overall structure relative to WT pol β is summarized on the correlation plot (right). I260Q data points deviating from the 1,1-cross-section display either altered structures (horizontal) or altered levels of enzymatic closure (vertical). Dots are colored according to the I260Q:DNA complexes indicated in the vector diagram at left