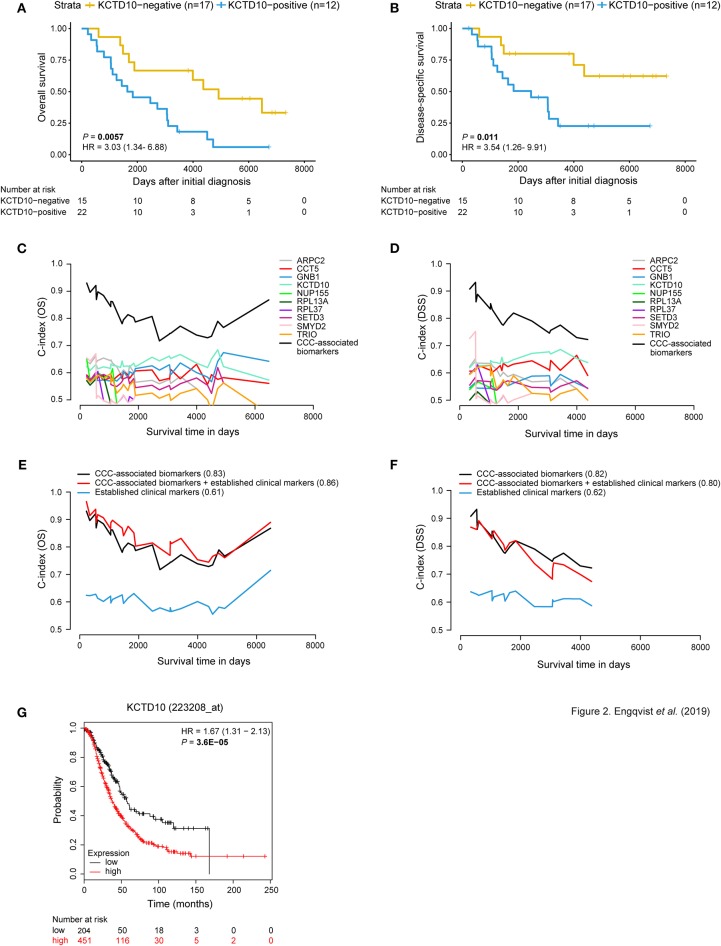
Figure 2.
Prognostic value of CCC-associated biomarkers. Kaplan-Meier survival plots (A,B) showing patient survival [overall survival (OS)/disease-specific survival (DSS)] in relation to dichotomized KCTD10 protein expression. Patients with KCTD10-positive staining (blue curve) correlated with both shorter OS and DSS [P-value = 0.0057, hazard ratio (HR) = 3.03 (95% confidence interval (CI) 1.34–6.88); P-value = 0.011, HR = 3.54 (95% CI 1.26–9.91)]. The x-axis depict days after initial diagnosis and the y-axis survival outcome (OS/DSS). The number of patients at risk by time (days after initial diagnosis) is shown below the Kaplan-Meier plot. Univariable and multivariable time-dependent area under the ROC curve [AUC(t)] plots (C,D) illustrating the predictive performance of each model over time and a significantly improved predictive model when combining the individual CCC-associated biomarkers (black curve) for OS and DSS. Multivariable survival plots for OS and DSS showing improved outcome prediction for CCC-associated biomarkers in comparison with established clinical markers (E,F). The outcome prediction concordance index (C-index) values are shown in parentheses. Further, the addition of protein expression status of CCC-associated biomarkers to established clinical markers resulted in improved outcome prediction (C-index = 0.86) for OS. Survival analysis was adjusted for age, stage, CA125, ploidy. The x-axis depicts survival time in days and the y-axis C-index values (OS/DSS). The prognostic value of each CCC-associated biomarker was validated using KM plotter for OS in HGSC and EC histotypes (n = 655) (G). Here, the prognostic value of KCTD10 is validated wherein patients with KCTD10-positive gene expression (patient samples with expression levels above the median) is shown in red and KCTD10-negative gene expression (patient samples with expression levels below the median) is shown in black. The number of patients at risk is indicated below the Kaplan-Meier plot. Cox proportional hazard models and log-rank tests were used to calculate HR, 95% confidence interval, and log rank P-value for Kaplan-Meier survival analysis and KM Plotter validation analysis.