Figure 2.
A Population of Apical Non-glial Progenitors Accounts for the Majority of Proliferating Progenitors in the Killifish Pallium
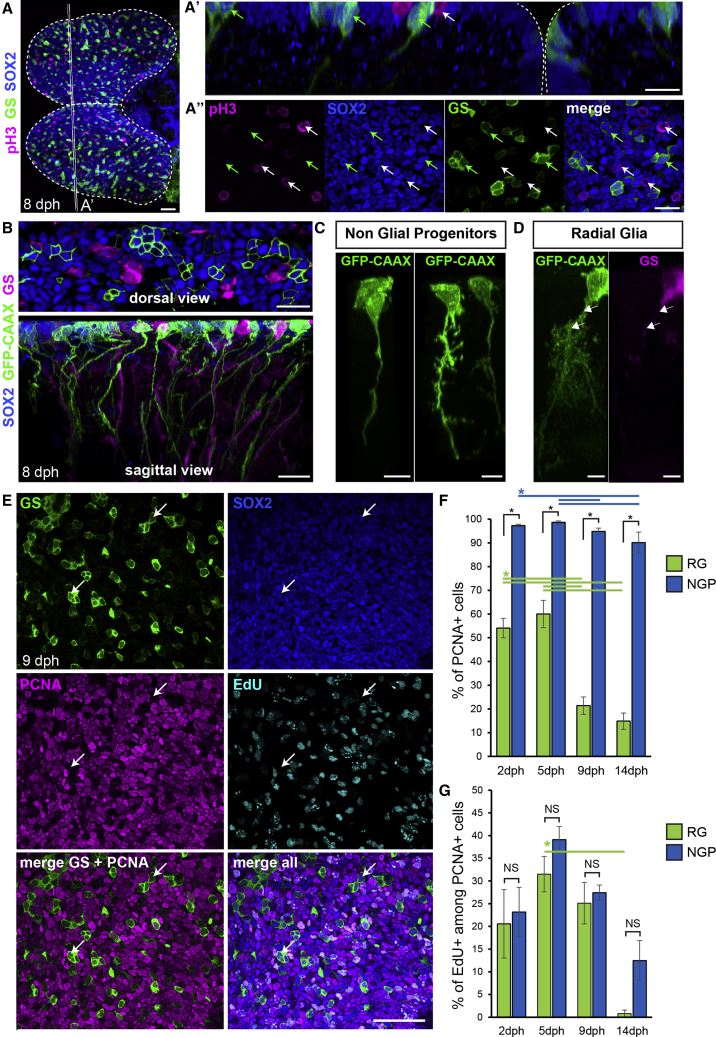
(A) Immunostaining for GS (green), Sox2 (blue), and pH3 (magenta) at 8 dph. Shown is a dorsal 3D view. Shown in (A’) is high magnification of a 5 μm transverse section through the 3D reconstruction. Shown in (A″) is high magnification of a single optical z-plane. White arrows point to non-glial NGP progenitors entering mitosis at the apical surface and green arrows point to sparsely distributed RGs.
(B) Shown at the top is the optical z-plane showing NGP electroporated with a GFP-CAAX-encoding plasmid (green), together with an immunostaining for GS (magenta) and Sox2 (blue). On the bottom is a sagittal view through a 3D reconstruction, highlighting the presence of a basal process on electroporated NGPs (green) and GS+ RGs (magenta).
(C) Examples of NGP cell morphology.
(D) Typical RG morphology, showing the ramified pattern of the basal process. Individual cells were manually detoured on a 3D reconstruction to visualize their morphologies.
(E) Optical z-plane showing immunostaining for GS (green), Sox2 (blue) and PCNA (magenta), and EdU detection (cyan), on the high magnification of the pallial surface at 9 dph. White arrows point to non-cycling RG cells.
(F) Proportion of PCNA+ cells among RGs (green bars) and NGPs (blue bars) at 2, 5, 9, and 14 dph.
(G) Proportion of EdU+ cells among PCNA+ cells among RGs (green bars) and NGPs (blue bars).
∗Corrected p value < 0.05; two-way ANOVA, followed by pairwise comparisons using Holm’s procedure. Data were rank transformed prior to analysis. Data are represented as mean ± SEM; n = 6, 5, 5, and 3 for 2, 5, 9, and 14 dph, respectively. Scale bars, 50 μm in (A) and (E); 20 μm in (A’), (A’’), and (B); and 10 μm in (C) and (D). See also Figure S2.