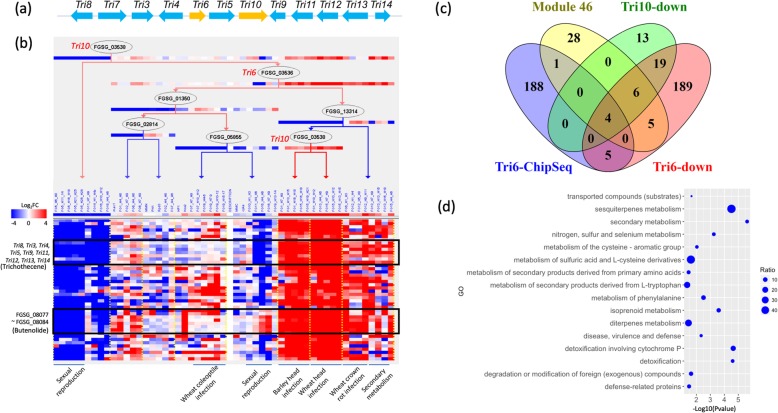
Fig. 3.
High accuracy of network inference highlighted by its capture of the trichothecene biosynthetic gene cluster. a. Diagram of the trichothecene biosynthetic gene cluster (Tri-cluster) in F. graminearum. b. Regulation program of the predicted module M46. The predicted regulators are arranged in a decision-tree structure that specifies the conditions under which the regulation occurs; these are delineated by the yellow vertical dotted lines. The heatmap underneath the tree shows the expression values of genes (rows) under different experimental conditions (columns). The experimental conditions are grouped into several major groups that are underlined; these are labeled at the bottom of the heatmap. The black empty box highlights all the Tri-cluster genes (labeled on the left) predicted in this module. c. Venn diagram displaying the overlap of M46 genes with the previously published down-regulated genes of ΔTri6 and ΔTri10 mutants as well as Tri6-binding genes identified by ChIP-seq. d. Dot plot showing the functional enrichment of M46 genes. The Y axis represents the significantly enriched GO terms. The X axis represents the P value (−Log10 transformed). The size of each dot is proportional to the gene ratio (Test/Reference), which was calculated as the number of genes in M46 (Test) divided by the number of genes in the F. graminearum genome (Reference) that are annotated with the GO term