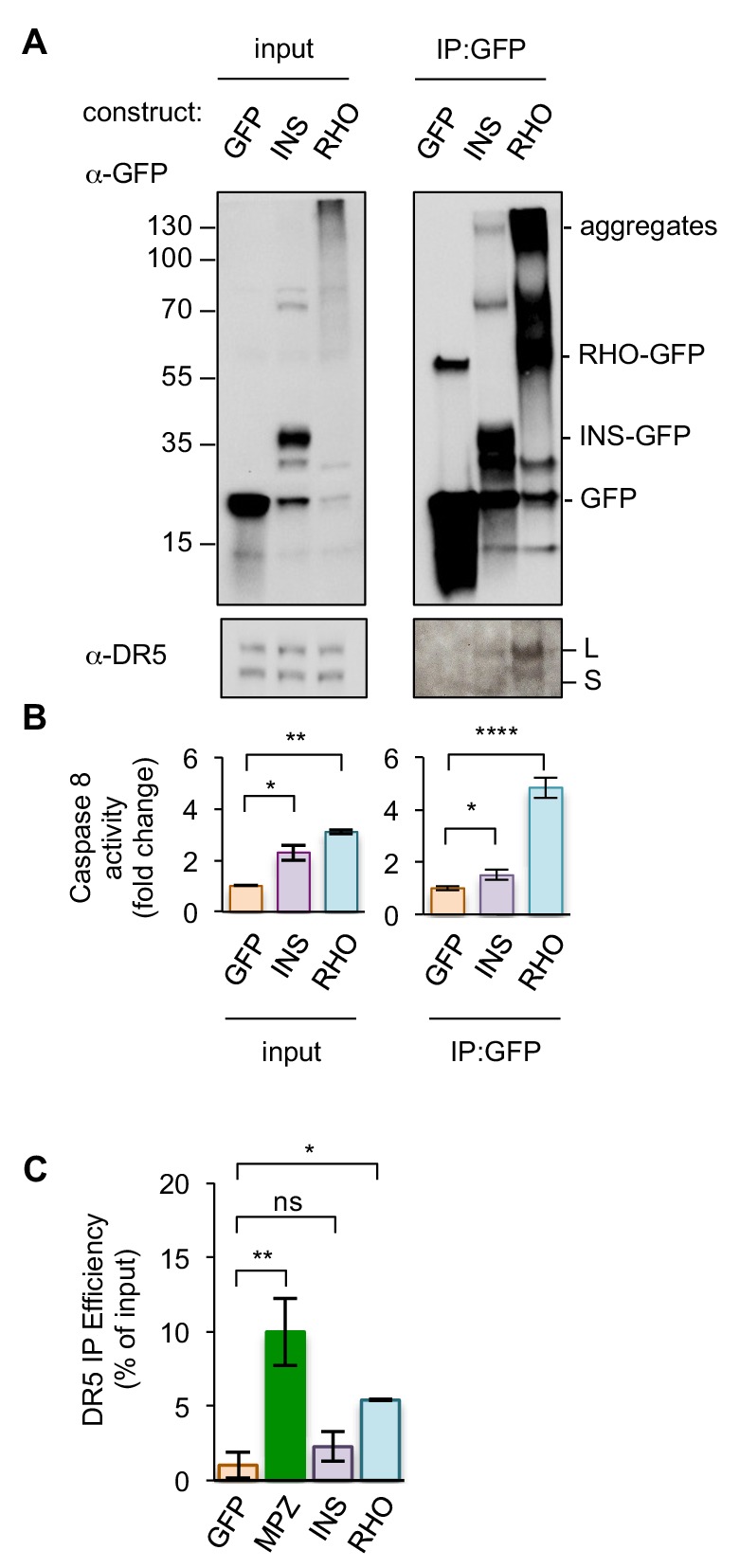
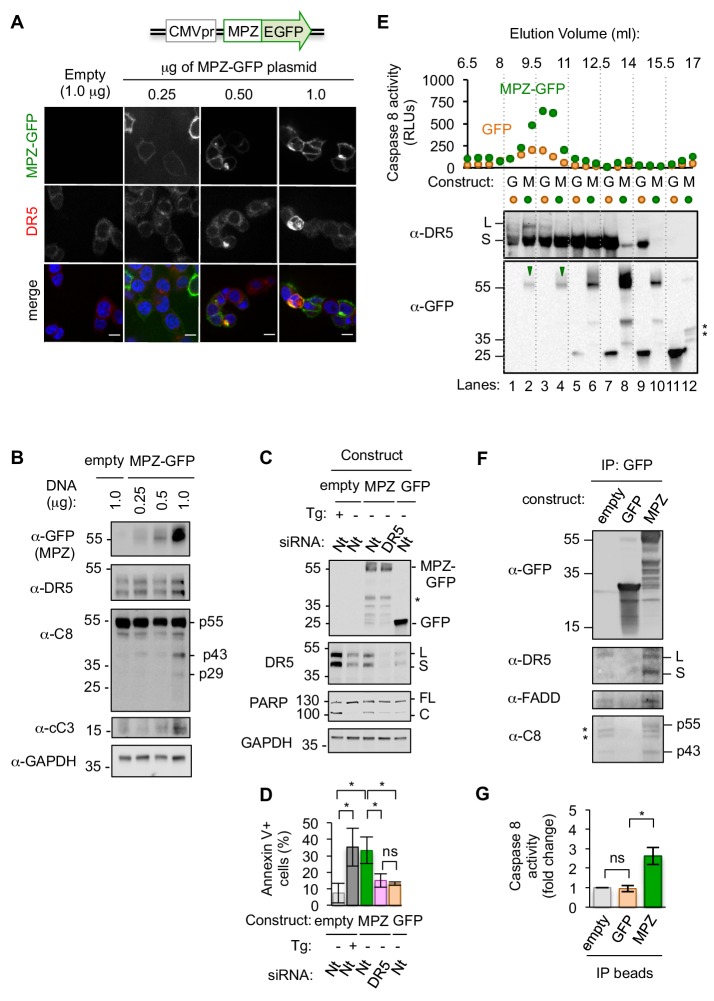
Figure 1. Misfolded proteins induce DR5-dependent apoptosis and assemble DR5-caspase 8 signaling complexes.
(A) Confocal images of epithelial cells HCT116 fixed 24 hr post-transfection with 0.25–1.0 μg of a plasmid containing myelin protein zero (MPZ) tagged with a C-terminal monomeric EGFP or 1.0 μg of the empty vector showing MPZ-GFP fluorescence (green) and immunofluorescence with an antibody against DR5 (red) (scale bar = 5 μm). (B) Western blot of HCT116 cell lysates harvested 24 hr post-transfection with a titration of MPZ-GFP plasmid or the empty vector (C8 = caspase 8, cC3 = cleaved caspase 3). p55 represents full-length, inactive C8; p43 indicates a C8 intermediate after release of the active p10 subunit, and p29 corresponds to the released p18 and p10 subunits. (C) Western blot of HCT116 cells transfected with siRNA against a non-targeting (Nt) control or DR5 (48 hr) followed by the empty vector -/+ 100 nM thapsigargin (Tg), 1.0 μg MPZ-GFP, or cytosolic GFP (24 hr; * denotes degradation products; L and S denote the long and short isoforms of DR5, respectively; FL and C denote full-length and cleaved PARP, respectively). (D) Average percent of annexin V staining for HCT116 cells transfected as described in C) from n = 3 biological replicates (error bars = SEM; * indicates p<0.05; ns indicates p=0.46 as analyzed by unpaired t-test with equal SD). See Figure 1—figure supplement 4D for gating. (E) Top: Caspase 8 activity in size exclusion chromatography fractions from lysates of HCT116 cells transfected with MPZ-GFP or cytosolic GFP (24 hr). Bottom: Size exclusion fractions were pooled according to dotted grid lines and immunoblotted for DR5 and GFP (* denotes degradation products). (F) Immunoprecipitation of GFP-tagged proteins from lysates of HCT116 transfected with MPZ-GFP, cytosolic GFP, or the empty vector (L and S denote the long and short isoforms of DR5, respectively). The percent of total DR5 recovered has been quantified in Figure 1—figure supplement 5C. (G) Fold change in caspase 8 activity relative to the empty vector control for beads with immunoprecipitated contents shown in Figure 1F (error bars = SEM for n = 3 biological replicates; * indicates p=0.023 and ns indicates p=0.83 as calculated by unpaired t-tests with equal SD).
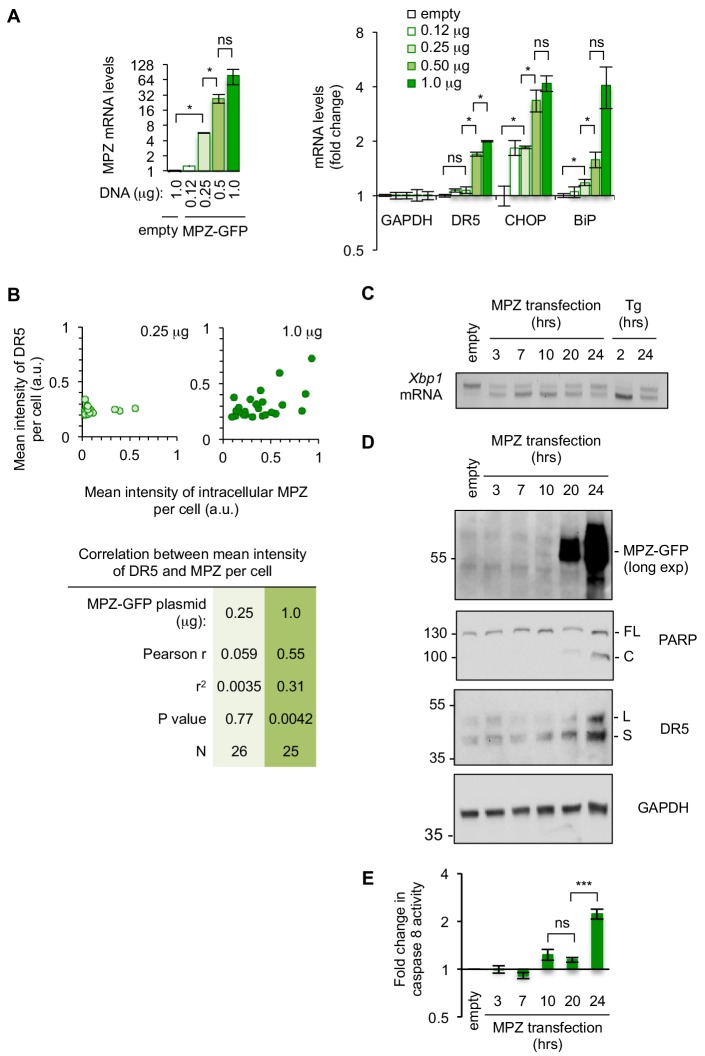
Figure 1—figure supplement 1. Sustained MPZ-GFP expression invokes a terminal, pro-apoptotic UPR at late time points.
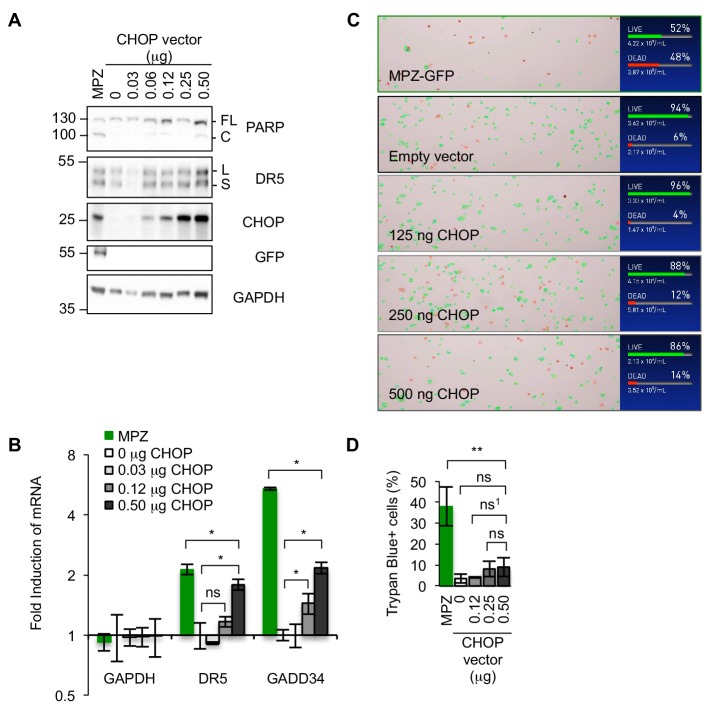
Figure 1—figure supplement 2. Upregulating DR5 levels in the absence of ER stress through ectopic expression of CHOP is not sufficient to induce apoptosis.
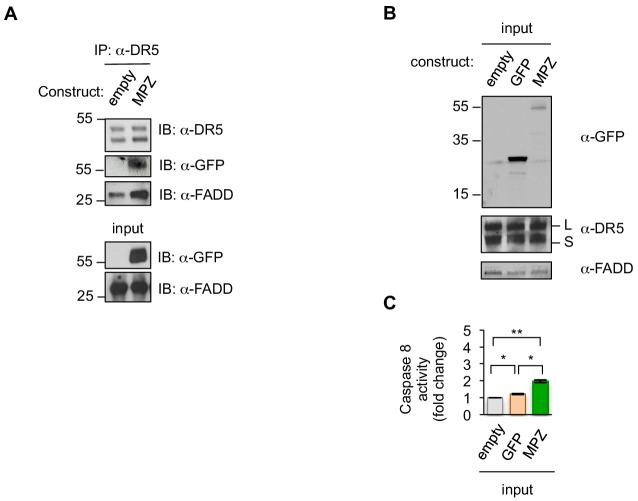
Figure 1—figure supplement 3. DR5 immunoprecipitates with FADD and MPZ-GFP.
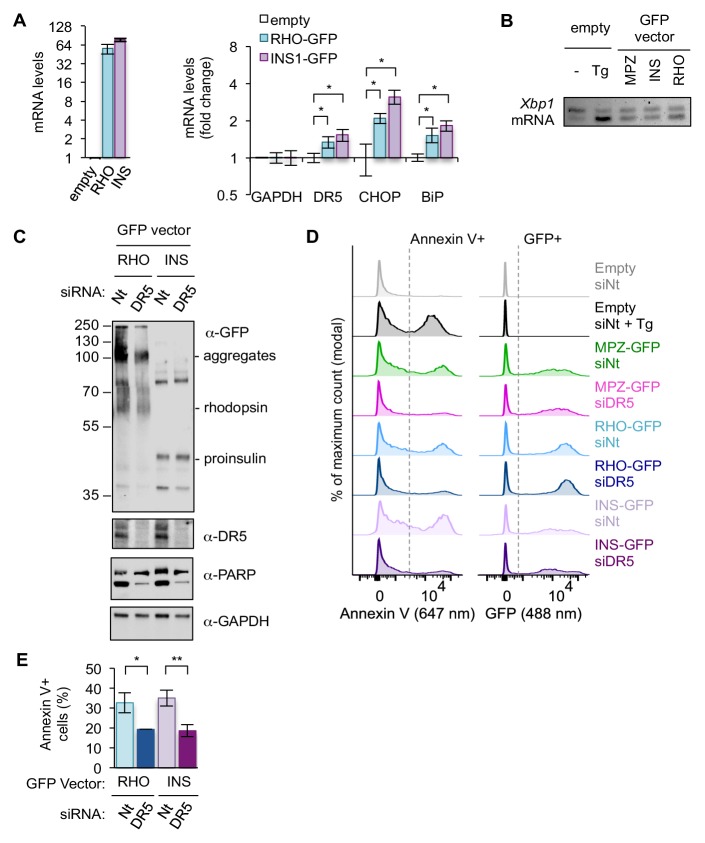
Figure 1—figure supplement 4. Sustained overexpression of other ER-trafficked proteins induce UPR-mediated apoptosis in a DR5-dependent manner.
Figure 1—figure supplement 5. DR5 engages a selective subset of ER-trafficked client proteins upon prolonged ER stress.