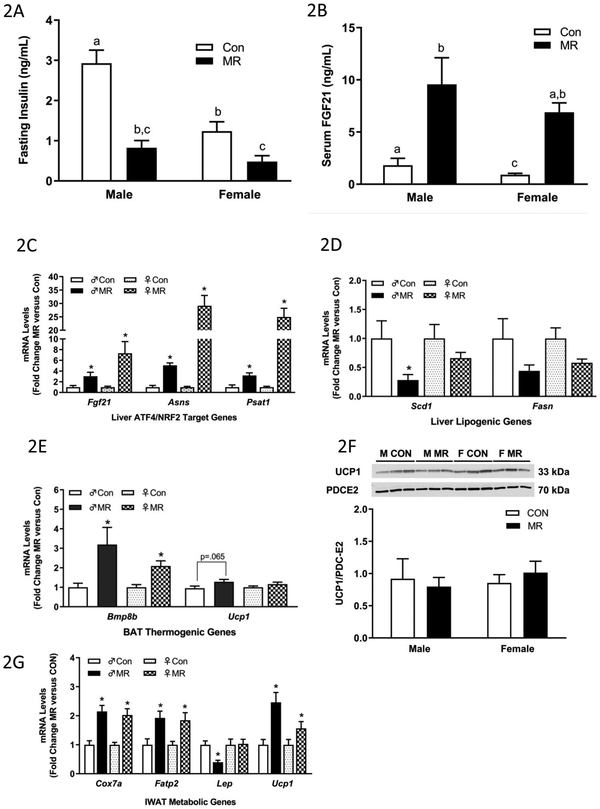
Figure 2 – Endocrine and Transcriptional Endpoints for Experiment 1.
Fasting insulin (A) and serum FGF21 levels (B) in blood samples collected at end of study. Hepatic gene expression (C,D), brown adipose tissue gene expression (E), and inguinal white adipose tissue (G) gene expression were measured via qPCR or by Western blot (F) for UCP1 expression in brown adipose tissue. The mitochondrial marker, PDC-E2 (pyruvate dehydrogenase dihydrolipoamide acetyltransferase) was used as a loading control for the UCP1 Western blot (E). Insulin and serum FGF21 were measured via ELISA. Con – control; MR – methionine restricted diets; IWAT – inguinal white adipose tissue; BAT – brown adipose tissue; Fgf21 – fibroblast growth factor 21; Asns – asparagine synthase; Psat1 - Phosphoserine Aminotransferase 1; Scd1 – stearoyl CoA desaturase 1; Fasn – fatty acid synthase; Bmp8b – bone morphogenetic protein 8b; Ucp1 – uncoupling protein 1; Cox7a – cytochrome C oxidase subunit 7A; Lep – Leptin; Fatp2 – fatty acid transport protein 2. Each variable was analyzed using a two way ANOVA with sex and diet as main effects and residual variance used as the error term. All values are expressed as mean ± SEM for 8 mice of each diet x sex. Means for fold-change of gene expression denoted with an asterisk (*) indicates significant difference from Con at p < 0.05. For fasting insulin and serum FGF21, group means not sharing a common letter differ at p < 0.05.