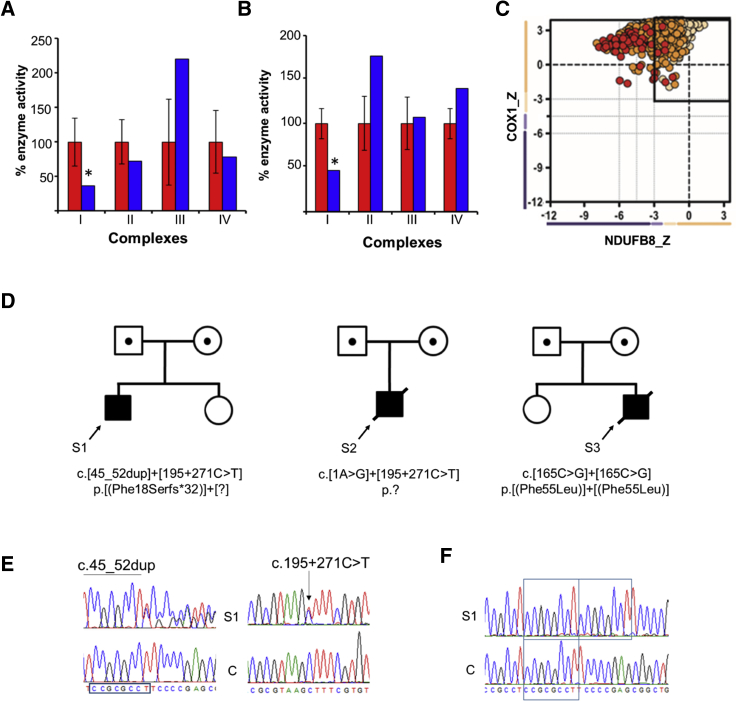
Figure 2.
Bi-Allelic NDUFAF8 Variants Are Identified in Three Unrelated Subjects
Respiratory chain enzyme analysis reveals a marked isolated complex I deficiency in subject 1’s skeletal muscle biopsy (shaded blue) compared to controls (shaded red) (A); this finding is recapitulated in the fibroblasts from subject 1 (B). Mean enzyme activities of muscle controls (n = 25) and fibroblast controls (n = 10) are set to 100%, with error bars representing standard deviation. The asterisk denotes a significant loss of enzyme activity.
(C) Quadruple immunofluorescent histochemical analysis of skeletal muscle biopsy from subject 1 demonstrates reduced levels of NDUFB8 (complex I) in the majority of single muscle fibers relative to those of the marker protein (porin), and expression of COX1 (complex IV) is preserved. Each dot represents a single muscle fiber. Black dashed lines represent the SD limits for the classification of the fibers. The X- and Y-axes represent the expression of NDUFB8 and COX-1: normal (< −1), intermediate +ve (−1 to −2 SD), intermediate −ve (−2 to −3 SD), and deficient (> −3 SD). The mean expression level of normal fibers is denoted a value of 0. Dots that fall outwith the solid box at -3 SD are strongly deficient fibres. Dots are color coded according to their mitochondrial mass (very low, blue; normal, beige; very high, red).
(D) Family pedigrees of subjects 1, 2, and 3 and corresponding recessive NDUFAF8 variants; compound heterozygous c.45_52dup (p.Phe18Serfs∗32) and c.195+271C>T (p.?) NDUFAF8 variants in subject 1; compound heterozygous c.1A>G (p.?) and c.195+271C>T (p.?) NDUFAF8 variants in subject 2 and a homozygous c.165C>G (p.Phe55Leu) NDUFAF8 variant in subject 3.
(E) Sequencing chromatograms depict the c.45_52dup and c.195+271C>T variants present in genomic DNA from S1.
(F) cDNA studies using RNA derived from subject 1 fibroblasts show that only the c.45_52dup allele is present at the mRNA level, supporting the possibility of degradation of the transcript associated with the c.195+271C > T variant.
S1 = subject 1; C = wild-type control. Variant nomenclature is according to GenBank accession NM_001086521.1