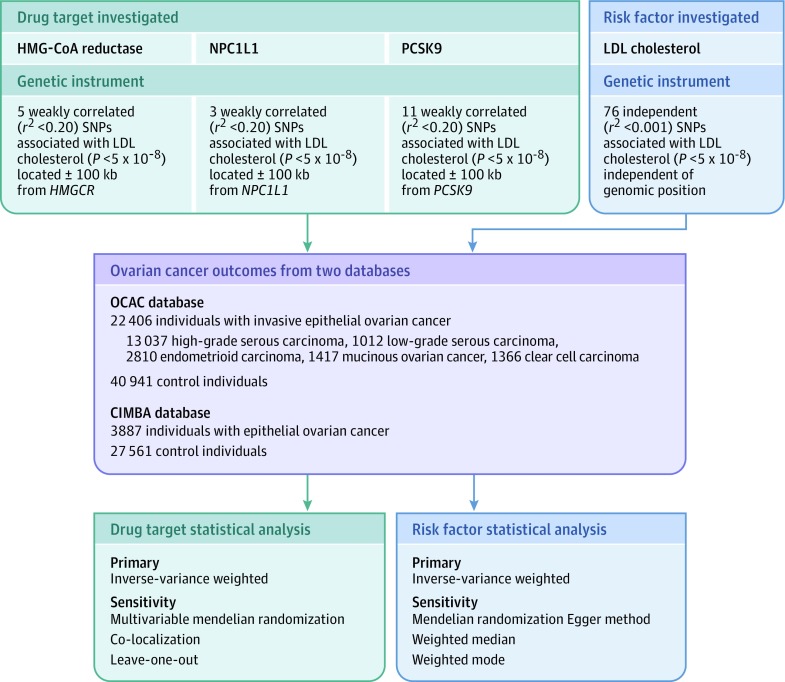
Figure 1. Genetic Instrument Construction, Data Sources, and Analysis Plan in a Study of the Association Between Genetically Proxied Inhibition of HMG-CoA Reductase and Epithelial Ovarian Cancer.
For each drug target or risk factor, genetic instruments were constructed by obtaining summary genetic association data on single-nucleotide polymorphisms (SNPs) associated with low-density lipoprotein (LDL) cholesterol located within or near the gene encoding the drug target (3-hydroxy-3-methylglutaryl coenzyme A [HMG-CoA] reductase, Niemann-Pick C1-Like 1 [NPC1L1], proprotein convertase subtilisin/kexin type 9 [PCSK9]) or independent of genomic position (LDL cholesterol) from Willer et al11 (N = 19 6465). Summary genetic association data for these SNPS were then extracted from genome-wide association studies of invasive epithelial ovarian cancer (Ovarian Cancer Association Consortium) and epithelial ovarian cancer in BRCA1/2 mutation carriers (Consortium of Investigators of Modifiers of BRCA1/2). After matching SNPs across data sets by assigning them the same effect allele, mendelian randomization analyses were performed using inverse-variance weighted random-effects models as primary analyses and various approaches as sensitivity analyses to test mendelian randomization assumptions (exchangeability and exclusion restriction). The bottom boxes represent the use of different sensitivity analyses to test mendelian randomization assumptions for drug target analyses and LDL cholesterol, because some of these sensitivity analyses require that variants used as genetic instruments are independent of each other, which was not the case for drug target analyses.