Figure 1.
The CoREST Complex Forms a Stable, Enzymatically Active, and Stoichiometric Complex
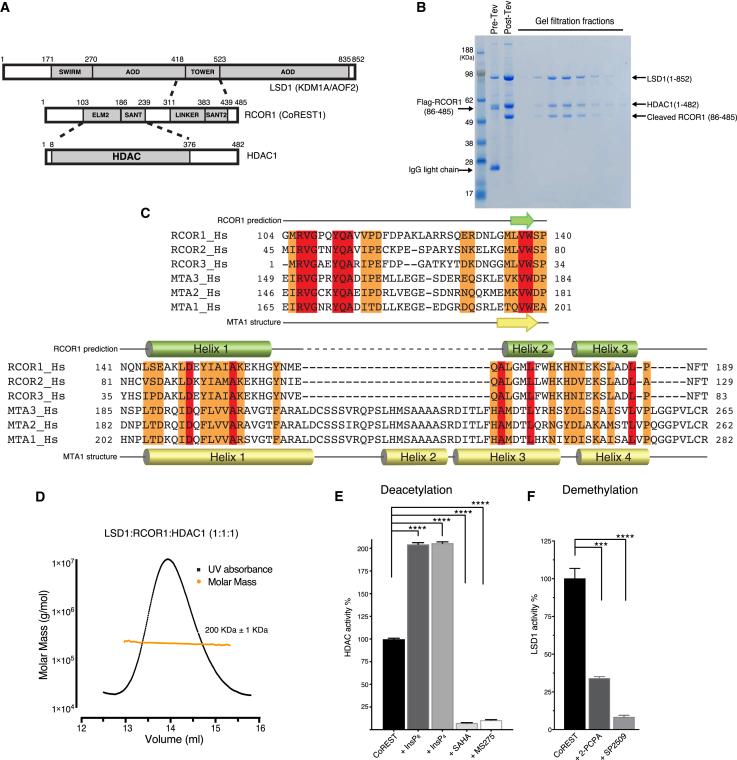
(A) Schematic representation of domain structures of LSD1/KDM1A/AOF2, CoREST/RCOR1, and HDAC1. Gray boxes represent the structured domains. Dashed lines indicate the interacting regions within the complex.
(B) Co-expression and purification of the LSD1:RCOR1:HDAC1 ternary complex.
(C) Sequence alignment of the ELM2 domain from RCOR1–3 and MTA1–3 proteins. Identical residues are shown in red, and conserved residues are shown in orange. The predicted secondary structure of RCOR1 is indicated above the sequence (green), and the secondary structure of MTA1 observed in the crystal structure is indicated below the sequence (yellow).
(D) Stoichiometry/molecular weight determination of the CoREST ternary complex by SEC-MALS.
(E) Deacetylase activity of the ternary complex. As expected, the activity is enhanced by 100 μM Ins(1,4,5,6)P4 (InsP4) and by Ins(1,2, 3,4,5,6)P6 (InsP6). The activity is inhibited by SAHA and MS275 (5 μM). The activity is normalized (100%) to the basal HDAC activity. The basal activity of the assay with no complex has been subtracted. Error bars indicate the SEM (n = 3). p values are shown in the form: ∗∗∗ p < 0.001 or ∗∗∗∗ p < 0.0001.
(F) Demethylase activity of the ternary complex. As expected, the activity is inhibited by 2-PCPA and SP2509 (10 μM). The activity is normalized (100%) to the basal demethylase activity. The basal activity of the assay with no complex has been subtracted. Error bars indicate the SEM (n = 3). p values are shown in the form: ∗∗∗ p < 0.001 or ∗∗∗∗ p < 0.0001.