Figure 4. Next-generation model for the molecular stratification of lung adenocarcinoma.
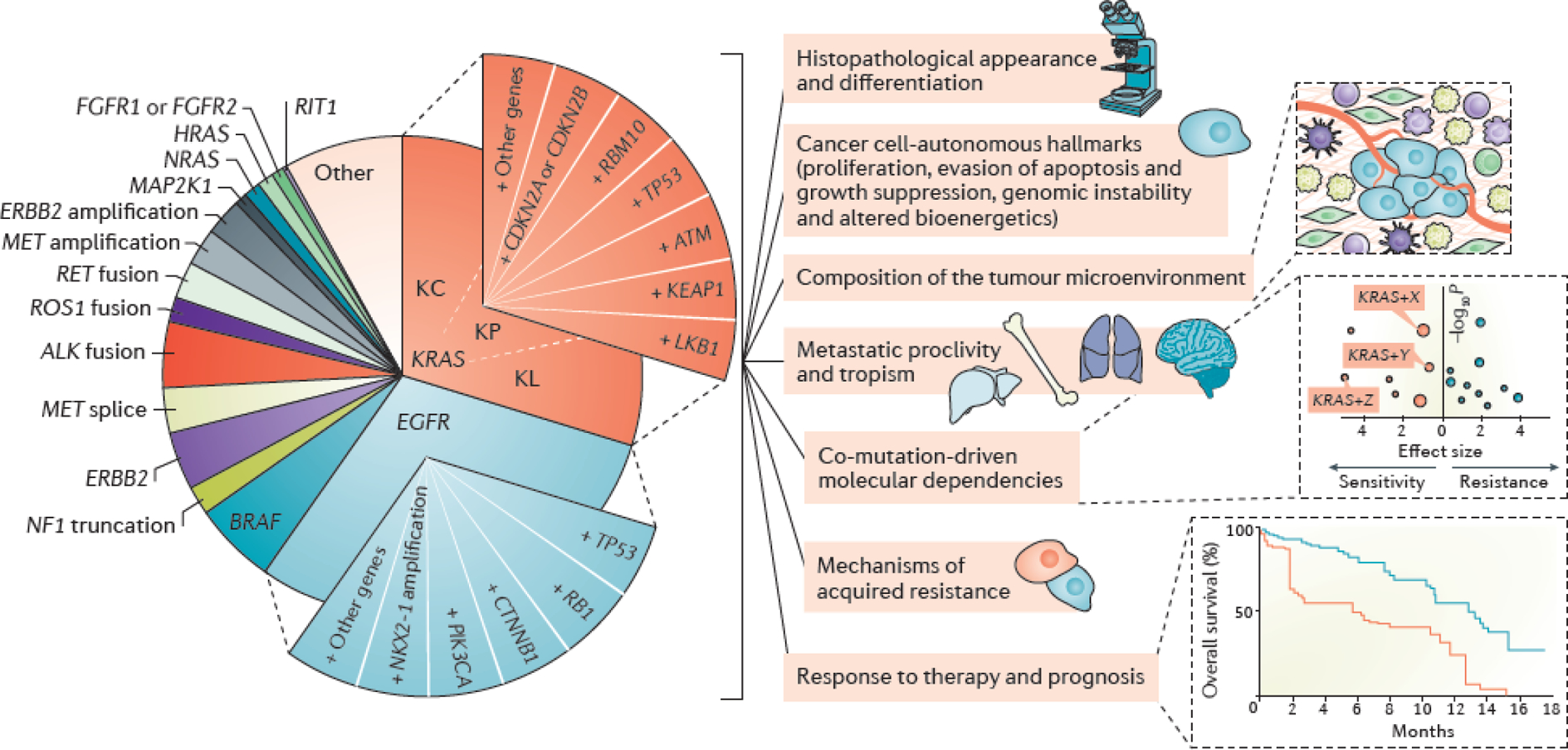
Oncogenic subgroups of lung adenocarcinoma (LUAD) are divided into smaller subsets on the basis of key co-occurring genomic alterations. Co-mutations constitute major determinants of tumor molecular diversity and can impact both tumor cell-autonomous and non-cell-autonomous cancer hallmarks; determine prognosis; predict response to systemic therapies and influence mechanisms of innate and acquired resistance. For simplicity, only KRAS and EGFR co-alterations are depicted graphically. For KRAS-mutant LUADs the previously identified KL, KP, and KC transcriptome-based subgroups are also indicated67; co-mutations in LKB1, KEAP1 and ATM are significantly enriched in the KL subgroup, whereas co-occurring alterations in TP53 and bi-allelic inactivation of CDKN2A/CDKN2B are hallmarks of the KP and KC subgroups respectively. Co-mutations in RBM10 don’t appear to exhibit predilection for any of the three KRAS transcriptomic subgroups. It should therefore be noted that several of the reported co-alterations within oncogene-defined groups are not mutually exclusive. Although co-mutation-defined cohorts are represented as slices of equal size, both the spectrum and prevalence of individual co-mutations evolve according to disease stage, prior treatment exposures, immune editing and the mutational processes that are operational at each stage of carcinogenesis.
