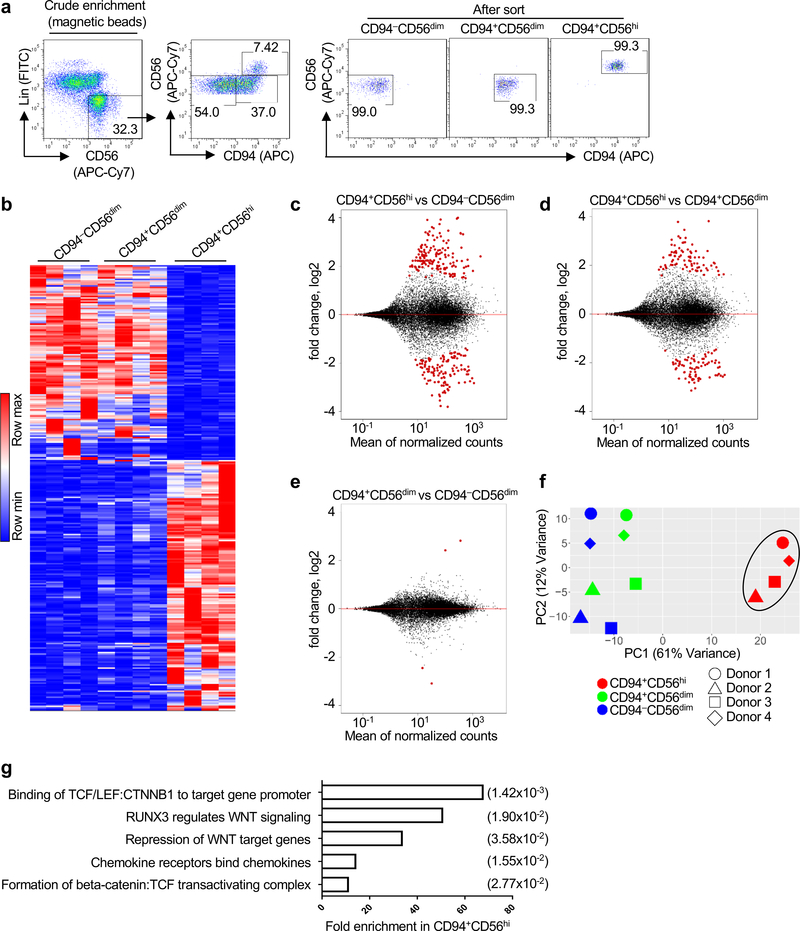
Fig. 4 |. TCF and WNT signaling pathway enrichment in CD94+CD56hiNK cells.
a, Sorting strategy for CD94−CD56dim, CD94+CD56dim, and CD94+CD56hi NK cell subsets.
b, Heatmap of differentially expressed genes by RNA-Seq (fold change of normalized counts, log2 >1, p<0.05 determined by DESeq2) for the indicated NK cell subsets sorted from four HIV-1− blood donors. c-e, Pairwise comparison of the indicated NK cell subsets based on differentially expressed genes. f, PCA based on RNA-Seq data from the indicated NK cell subsets. g, Reactome pathway analysis based on 152 differentially expressed genes in CD94+CD56hi NK cells. GENEONTOLOGY produces p value from Fisher’s exact test, followed by BH false discovery rate (in parentheses). All data were generated using blood from HIV-1− donors.