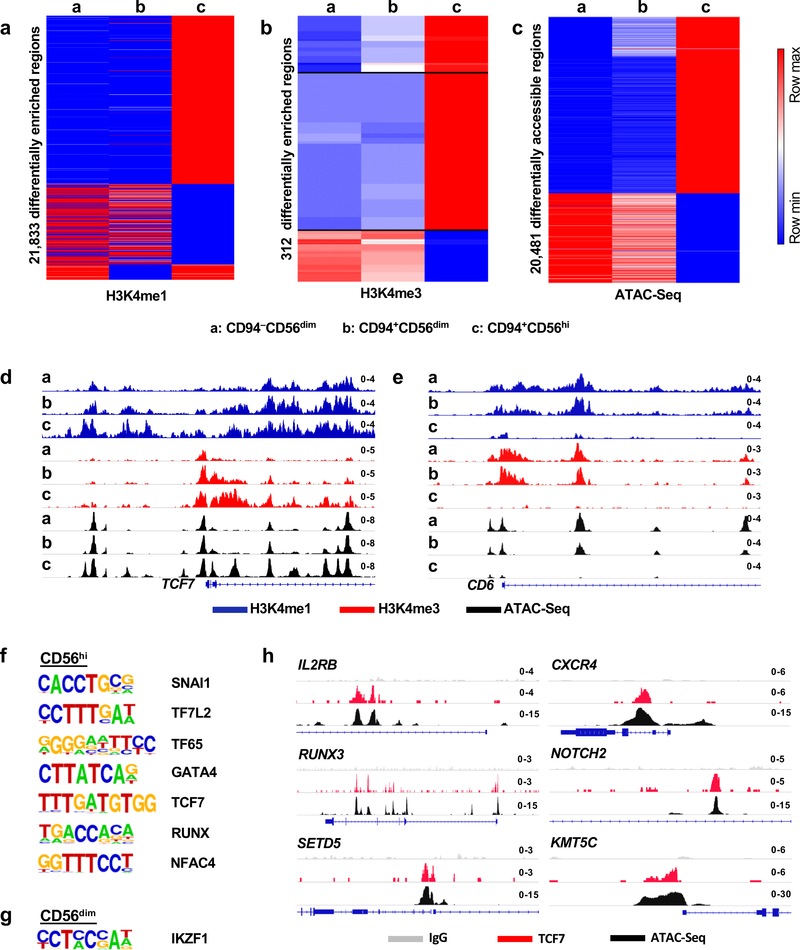
Fig. 5 |. Distinct chromatin landscape in CD94+CD56hi NK cells.
a-c, Heatmap showing differential enrichment (>2 fold change of normalized counts) for H3K4me1 (a) and H3K4me3 (b) by CUT&RUN, or accessible chromatin by ATAC-Seq (c), in sorted CD94−CD56dim, CD94+CD56dim, and CD94+CD56hi NK cell subsets. Data is representative of two HIV-1− blood donors. d,e, H3K4me1, H3K4me3, and ATAC-Seq signal on TCF7 (d) and CD6 (e), in the indicated NK cell subsets. f,g, De novo analysis of transcription factor binding motifs enriched in open chromatin from CD56hi NK cells (f), or CD56dim NK cells (g), using HOMER. h, ATAC-Seq and TCF7 CUT&RUN signal at the indicated loci from the three NK cell subsets. All data were generated using blood from HIV-1− anonymous donors.