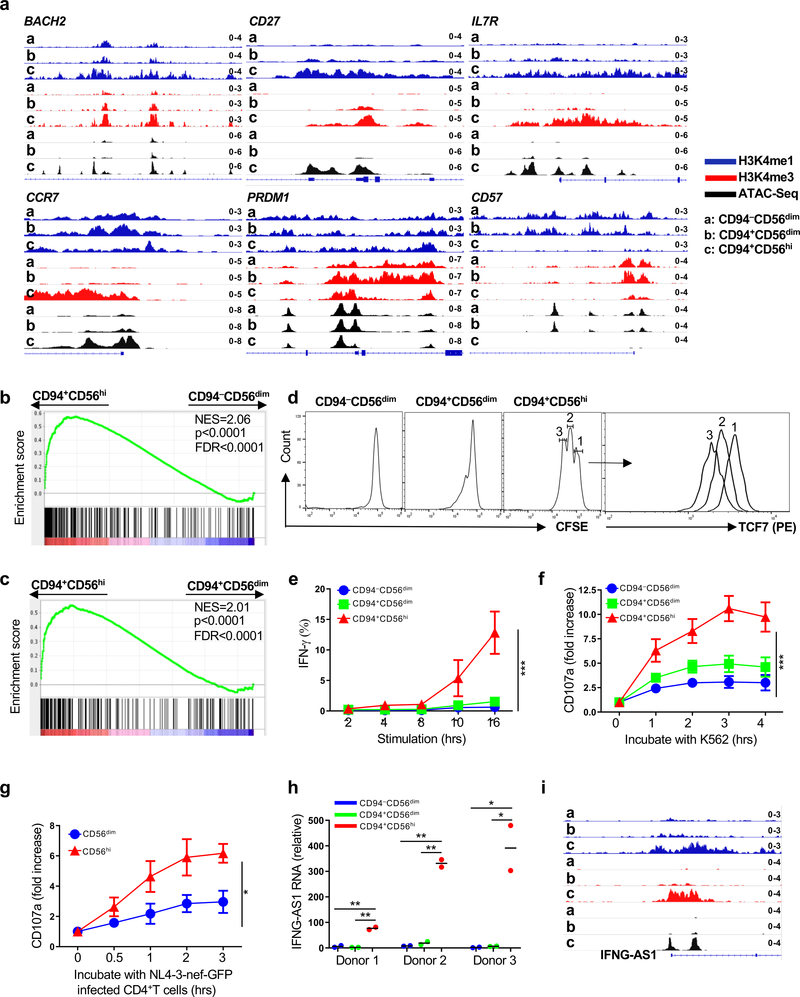
Fig. 6 |. Chromatin, transcriptome, and phenotype define CD94+CD56hi NK cells as memory cells.
a, CUT&RUN and ATAC-Seq showing memory gene-associated loci in sorted CD94−CD56dim, CD94+CD56dim, and CD94+CD56hi NK cells. b,c, Gene set enrichment analysis comparing memory CD8+ T cell expression signature (GSE9650) with sorted CD94+CD56hi and CD94−CD56dim NK cells (b), or with sorted CD94+CD56hi and CD94+CD56dim NK cells (c). p value and FDR determined using GSEAPreranked module from GenePattern with 1000 permutations. d, Sorted NK cells labelled with CFSE and cultured 5 days in IL-15 (5 ng/ml). CFSE and TCF7 detected by flow cytometry. e, Percent IFN-γ+ cells by flow cytometry after stimulation of sorted NK cell populations with IL-12 and IL-15 (n=4). f, Fold increase of surface CD107a on bead-enriched NK cells after incubation with K562 cells (n=4). g, bead-enriched NK cells incubated with IL-15 (5ng/ml) for 5 days, then with NL4-3-nef-GFP infected CD4+ T cells. Fold increase of surface CD107a on indicated NK populations, as in f (0h, n=5; 0.5h, 1h, 2h, n=3; 3h, n=5). h, IFNG-AS1 expression relative to GAPDH by RT-qPCR for the indicated NK cell populations sorted from 3 donors. Each population contains 2 technical replicates. i, IFNG-AS1 chromatin analysis as in a. Data are mean ± s.e.m. e-g, two-way ANOVA; h, two-tailed unpaired t-test. *p<0.05, **p<0.01, ***p<0.001. All data were generated using blood from HIV-1− anonymous donors.