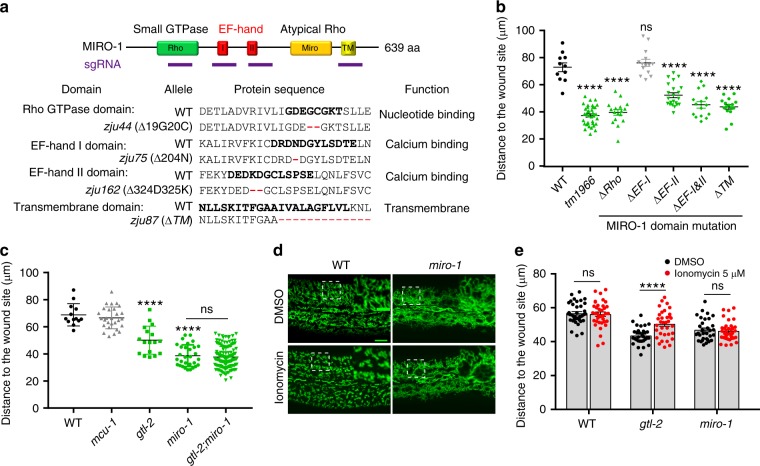
Fig. 4. WIMF is dependent on wound-induced Ca2+-MIRO-1 signaling.
a Top, experimental design to disrupt the function of MIRO-1 protein domains using CRISPR-Cas9 mediated mutagenesis. Bottom, miro-1 in-frame mutation alleles. Sequences in bold are key residues in each domain40. Details of indel mutations are in Supplementary Table 5. b Quantitation of the distance of the fragmented mitochondria to the injury site shown on the left in different animals (WT, n = 11; tm1966, n = 30; ΔRho, n = 19; ΔEF-I, n = 14; ΔEF-II, n = 22; ΔEF-I&II, n = 15; ΔTM, n = 16 animals). Note, mutations of the Rho, EF-hand II and TM domains show reduced spreading of WIMF compared to the WT and similar to miro-1(tm1966) animal, indicating that these mutations affect MIRO-1’s function. Bars indicate mean ± SEM. ns, P = 0.8321, ****P < 0.0001 (versus WT), One-way ANOVA Dunnett’s test. c Quantitation of the distance of the fragmented mitochondria to the injury site in TRPM/gtl-2(n2618), mcu-1(ju1154), and gtl-2(n2618);miro-1(zju19) mutant (WT, n = 13; gtl-2, n = 17; miro-1, n = 36; gtl-2;miro-1, n = 143 animals). miro-1(zju19) and miro-1(zju40) frameshift mutations were generated by CRISPR-Cas9 mediated mutagenesis in gtl-2 background. Bars indicate mean ± SEM. ns, P = 0.9558 (versus miro-1), two-tailed unpaired t-test; ns, P = 0.9558, ****P < 0.0001 (versus WT), One-way ANOVA Dunnett’s test. d Representative confocal image of mitochondrial morphology after treatment with Ca2+ ionophore ionomycin in both WT and miro-1 mutants. Scale bars, 10 μm, and 5 μm (zoom-in). N = 3 independent experiments. e Quantitation of the distance of the fragmented mitochondria to the injury site in gtl-2(n2618), miro-1(tm1966) mutant treated with low concentration of ionomycin (2.5 μΜ treated for 1 h) (WT-DMSO, n = 37; WT-ionomycin, n = 36; gtl-2-DMSO, n = 31; gtl-2-ionomycin, n = 34; miro-1-DMSO, n = 32; miro-1-ionomycin, n = 35). Bars indicate mean ± SEM. ns, P = 0.7435 (WT) or 0.6382 (miro-1), ***P < 0.0001, Two-tailed unpaired t-test. Source data are provided as a Source Data file.