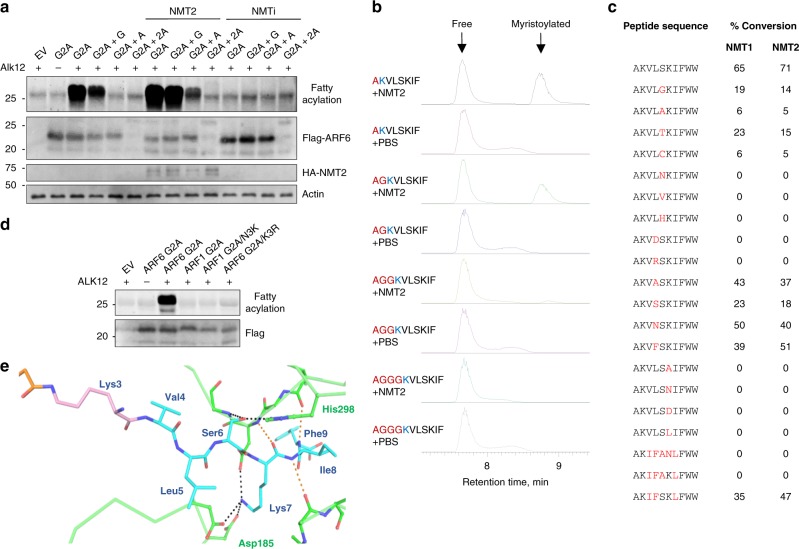
Fig. 3. Sequence requirement for NMT lysine myristoylation.
a Cellular Alk12 labeling results with the indicated mutants showing that NMT can accommodate other positions of K at the N-terminus in cells. The mutant containing two additional alanine residues at the N-terminus could not be stably expressed. b NMT can accommodate other N-terminal positions of K in vitro on synthetic peptides. Ion chromatograms searched for the substrate and product ions are shown. c Amino acid sequence preference for NMT lysine myristoylation on ARF6 G2A-derived peptides. Red letters represent changed residues. The sequence IFANL is the ARF1-derived sequence. The product was detected by LC-MS and the percentage of conversion was calculated from the substrate and product peak areas on HPLC UV traces. d Alk12 labeling of the indicated mutants transiently overexpressed in HEK293T cells showing that, unlike ARF6 G2A, ARF1 G2A/N3K is not myristoylated. e Peptide recognition for Lys3 myristoylation appears similar to that for Gly2 myristoylation. NMT2 residues are shown in green. Hydrogen bonding to Ser6 by His298 and Gly472 backbone nitrogen and to Lys7 side chain by a cluster of aspartates 183, 185, and 471 are shown as black dotted lines. Further hydrogen bonding to the backbone of the peptide (orange dotted lines) is also the same as in previous structures.