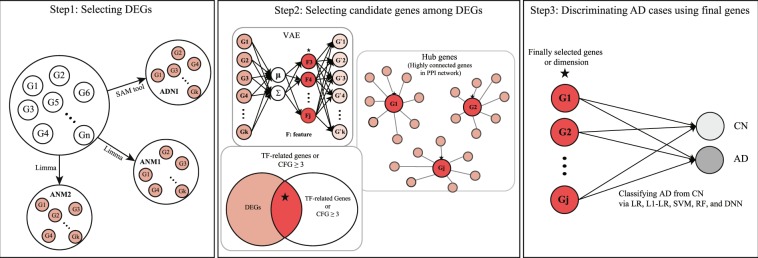
Figure 1.
A framework of the study. The informative genes were selected using a training set by two processes: (Step 1) Extracting DEGs; (Step 2) Selecting informative genes using feature selection methods, including VAE, TF-related genes, hub genes, and the CFG scoring; (Step 3) Learning a prediction model using a training set, and predicting a test set by employing five classification methods, including logistic regression (LR), L1-regularized LR (L1-LR), SVM, RF, and DNN. Stars denote the best performance among five classifying methods. DEG, differentially expressed gene; SAM, significance analysis of microarray; ADNI, Alzheimer’s Disease Neuroimaging Initiative; ANM, AddNeuroMed; VAE, variational autoencoder; TF, transcription factor; PPI, protein-protein interaction; CFG, Convergent Functional Genomics; LR, logistic regression; SVM, support vector machine; RF, random forest; DNN, deep neural network; CN, healthy control; AD, Alzheimer’s disease.