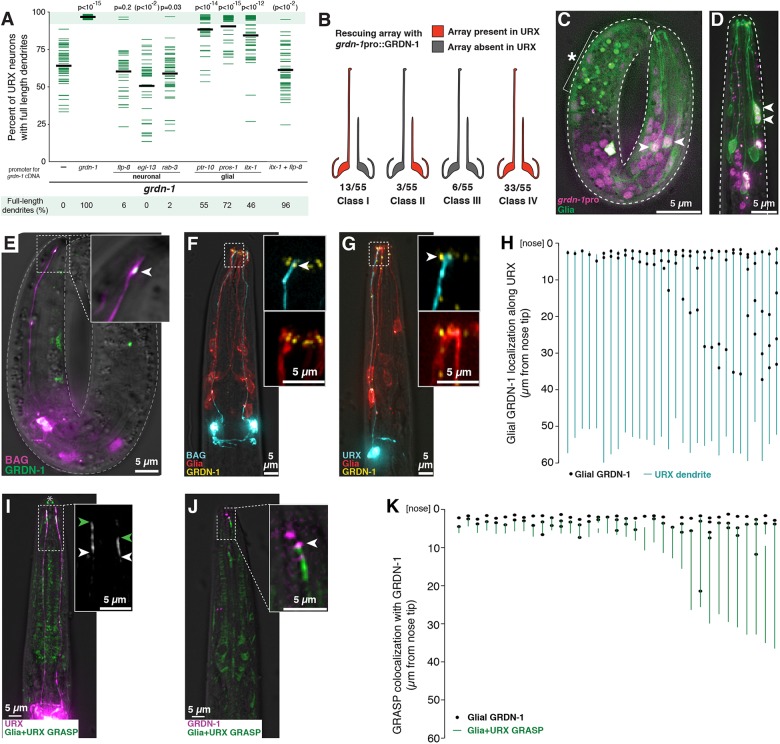
Fig. 7.
GRDN-1 can act in glia and localizes in puncta at dendrite contacts. (A) Transgenes containing GRDN-1a cDNA under control of the indicated promoters, or no transgene (−, same data as Fig. 3), were introduced into grdn-1 animals and URX dendrite lengths were measured as a percentage of the distance from the cell body to the nose. P-values (Wilcoxon Rank Sum test) compared with grdn-1 with no transgene (−) are shown at the top. Parentheses indicate that, for unknown reasons, expression using egl-13pro enhances the defects. Colored bars represent individual dendrites (n≥34 per genotype); black bars represent population averages. Shaded region represents wild-type mean±5 s.d. and the percentage of dendrites in this range (‘full-length dendrites’) is indicated below the plots. Data for BAG are in Fig. S8. (B) Mosaic analysis was performed using grdn-1 animals bearing a stably integrated URX marker (flp-8pro:GFP) to assess dendrite length and an extrachromosomal transgene, which is subject to stochastic loss during cell division, containing grdn-1pro:GRDN-1a and the URX marker flp-8pro:CFP to assess the ability of GRDN-1 to act cell-autonomously. Animals in which only the left or right URX dendrite was full-length were selected as probable mosaics, and were scored for the presence (red) or absence (gray) of the rescuing array in each URX neuron. (C,D) Wild-type embryo (C) and L1 stage larva (D) expressing a histone-mCherry fusion protein under control of the grdn-1 promoter (grdn-1pro:his-24-mCherry, magenta) and a membrane-localized GFP to mark glia (itx-1pro:myristyl-GFP, green). Arrowheads indicate examples of cells expressing both markers. Asterisk and bracket indicate background auto-fluorescence due to gut granules. (E-G) The localization of glial-expressed GRDN-1a relative to URX and BAG was visualized (E) in embryos, by expressing sfGFP-GRDN-1a in glia (itx-1pro, green) together with an embryonic BAG marker (egl-13pro:mCherry, purple); and (F,G) in L1 stage larvae, by expressing YFP-GRDN-1a (itx-1pro, yellow) and myristyl-mCherry (itx-1pro, red) in glia together with CFP (blue) in BAG (F, flp-17pro) or URX (G, flp-8pro). Arrowheads indicate glial-expressed puncta of GRDN-1a near dendrite endings. (H) The positions of glial-expressed GRDN-1a puncta were quantified relative to the URX dendrite. Each line represents a single URX dendrite and adjacent YFP-GRDN-1a puncta are shown as black dots. n=30. (I,J) GRDN-1a puncta were visualized relative to the dendrite-glia contact. (I) Regions of direct dendrite-glia contact were visualized using a GFP-reconstitution assay (GRASP) targeting URX and glia (itx-1pro:CD4-spGFP1-10 and flp-8pro:CD4-spGFP11, green). URX is also marked by mCherry (flp-8pro, magenta). Green and white arrowheads mark the anterior and posterior extents of the GRASP signal. Owing to the faint signal, prominent autofluorescence at the mouth (asterisk) and elsewhere in the head is visible. (J) The localization of glial-expressed GRDN-1a relative to the URX-glia contact was visualized by expressing mApple-GRDN-1a in glia (itx-1pro, magenta) together with GRASP (green). Arrowhead indicates glial-expressed punctum of GRDN-1a at the anterior extent of the dendrite-glia contact revealed by GRASP. (K) The positions of glial-expressed GRDN-1a puncta were quantified relative to the dendrite-glia contact. Each line represents one URX-glia pair, as revealed by GRASP, and adjacent mApple-GRDN-1a puncta are shown as black dots. Multiple dots may reflect expression of the promoter in several (∼6-12) glial cells. n=35.