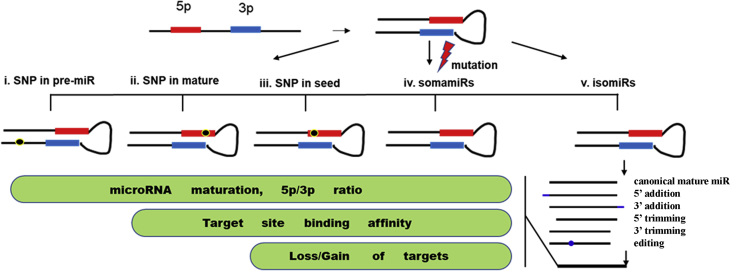
Fig. 1.
Diagrammatic summary of single nucleotide variations (SNVs) in microRNAs. SNVs can be classified into three categories: single nucleotide polymorphisms (SNPs), somatic mutations (somamiRs) and isomiRs. SNPs in pre-miRs, mature miRs or within the seed sequences may (1) influence the biogenesis/maturation of the mature miRs and cause change in 5p/3p ratios, (2) strengthen or reduce the binding affinity for the mature miRs to bind to their targets or (3) change miRs' targetome through loss or gain of binding sites. SomamiRs, which are somatically mutated microRNAs, could be considered as a combination of all possible SNPs. IsomiRs are bona fide microRNA variants or isoforms that result from either addition, depletion/trimming or microRNA editing and usually show race, gender, population, and disease subtype dependencies. All of these SNVs modulate the intracellular microRNA-mRNA interaction network.