Abstract
Background
Intervertebral disc degeneration (IDD) is a major contributor to lower back pain, however, the molecular and pathogenetic mechanisms underlying IDD are poorly understood. As a high-risk factor for IDD, compression stress was reported to induce apoptosis of nucleus pulposus (NP) cells and extracellular matrix (ECM) degradation during IDD progression. Circular RNA (circRNA) is a class of endogenous non-coding RNA (ncRNA) and has been reported to function in several diseases. However, whether and how circRNA regulates compression-induced damage of NP cells remains vague. Here, we aimed to investigate the key role of circRNA in compression loading-induced IDD.
Methods
We analysed the circRNA expression of three samples from compression-treated NP cells and three control samples using circRNA microarray assays and further investigated the circRNA involved in compression-induced damage of NP cells (circRNA-CIDN). We investigated the effects of circRNA-CIDN on compression-induced cell apoptosis and NP ECM degradation in vitro and ex vivo. We observed that circRNA-CIDN bound to miRNAs as a miRNA sponge based on luciferase and RNA immunoprecipitation (RIP) assays.
Findings:
CircRNA-CIDN was significantly downregulated in compression-treated human NP cells, as validated by circRNA microarray and qRT-PCR analysis, and overexpressing circRNA-CIDN inhibited compression-induced apoptosis and NP ECM degradation. Further studies demonstrated that circRNA-CIDN served as a sponge for miR-34a-5p, an important miRNA that enhanced compression-induced damage of NP cells via repressing the silent mating type information regulation 2 homolog 1 (SIRT1). CircRNA-CIDN was also verified to contain IDD development in an ex vivo IDD model.
Interpretation
Our results revealed that circRNA-CIDN binding to miR-34a-5p played an important role in mitigating compression loading-induced nucleus pulposus cell damage via targeting SIRT1, providing a potential therapeutic strategy for IDD treatment.
Funding
National Natural Science Foundation of China (81772391, 81974348), Fundamental Research Funds for the Central Universities (2017KFYXJJ248).
Keywords: Circular RNAs, miR-34a-5p/SIRT1 axis, Compression, Intervertebral disc degeneration, Nucleus pulposus cell
Research in context.
Evidence before this study
Lower back pain (LBP) is one of the most common chronic pain conditions which has caused an enormous socio-economic burden worldwide. Intervertebral disc degeneration (IDD) is regarded as the leading cause of lower back pain. Previous research has found that compression stress was a high-risk factor for IDD and it could induce apoptosis of nucleus pulposus (NP) cells and extracellular matrix (ECM) degradation to accelerate IDD. Accumulating evidence demonstrated that a new class of endogenous non-coding RNA named circular RNA (circRNA) played important roles in several diseases recently. However, there are no researches investigating whether and how circRNA regulates compression-induced damage of NP cells during IDD.
Added value of this study
Using circRNA microarray assays and further analysis, circRNA-CIDN was characterized and it was significantly downregulated in compression-treated human NP cells compared to that in the normal samples. Overexpressing circRNA-CIDN inhibited compression-induced cell apoptosis and NP ECM degradation. Further studies demonstrated that circRNA-CIDN served as a sponge for miR-34a-5p, an important miRNA that enhanced compression-induced damage of NP cells via repressing SIRT1. We also verified that circRNA-CIDN was able to contain IDD development in an ex vivo IDD model. Here, we provided the first evidence for the effects of circRNA on compression loading-induced damage in human NP cells during IDD progression.
Implications of all the available evidence
This study revealed that circRNA-CIDN binding to miR-34a-5p played an important role in mitigating compression loading-induced nucleus pulposus cell damage via targeting SIRT1. The circRNA-CIDN/miR-34a-5p/SIRT1 axis provides a potential therapeutic strategy for the clinical treatment of IDD.
Alt-text: Unlabelled box
1. Introduction
Lower back pain (LBP) is one of the most common chronic pain conditions, and over 80% of adults will suffer from LBP at a certain age, and this has caused an enormous socio-economic burden worldwide [1]. Intervertebral disc (IVD) degeneration (IDD) has been regarded as the primary underlying cause of LBP [2]. However, apart from limited treatments such as surgery, pharmacological, and nonpharmacological approaches to relieve pain, there are currently no effective and constructive therapeutic strategies for treating IDD, since its pathogenesis has not been explicitly expounded yet [3]. Therefore, it will be of great significance to reveal the molecular and pathogenetic mechanisms underlying IDD.
The intervertebral disc is the biggest avascular organ in the body and consists of a central gelatinous nucleus pulposus (NP) and a surrounding annulus fibrosus (AF) [4]. The NP cells are the main resident cells in the highly hydrated NP tissue and are responsible for controlling NP extracellular matrix (ECM) synthesis and decomposition, maintaining the normal structural and functional properties of the IVD [5]. When the NP ECM catabolism exceeds anabolism within IVD, aggrecan and collagen II are degraded, resulting in NP dehydration and resorption, decrease of disc height, and decline of ability to resist mechanical stress and this is one of the major pathological characteristics of degenerative disc diseases [6,7]. Matrix-degrading matrix metalloproteinases (MMP) such as MMP-3 and MMP-13 are important ECM-degrading enzymes during this process [8]. Another pathological feature of IDD is abnormally elevated NP cell apoptosis levels, which is accompanied by cleaved caspase-3 and Bax expression up-regulation and Bcl-2 protein down-regulation [6].
As a load absorbing and buffering unit of the spine, the IVD is subjected to various degrees of mechanical stress in daily life. The pressure of the IVD has been measured to range from 0.1 MPa when lying face down to 2.3 MPa when lifting a 20 kg weight with a rounded and flexed back [9,10]. Also, the pressure of human L4–L5 IVD was measured as 0.5 MPa when standing relaxed and 1.1 MPa when standing flexed forward, as reported by Wilke et al. [11]. Studies revealed that spinal mechanical stress could increase the risk of IDD among general population [12]. Mechanical loading was thought to be primarily responsible for remodeling the intervertebral disc, which may result in disc injury over time [13,14]. Furthermore, our previous research, among others in literature, suggested that excessive loading induced apoptosis of NP cells and ECM degradation, by which it was deeply involved in IDD progression [14], [15], [16].
While a variety of factors have been shown to be involved in the pathophysiological mechanisms of IDD, genetics is thought to be one of the most critical contributors, and non-coding RNAs (ncRNAs) have been increasingly reported to play a key role during the process of IDD [17,18]. MicroRNAs (miRNAs) are small, highly conserved endogenous ncRNAs, and most of them regulate gene expression post-transcriptionally in mammals by guiding the RNA-induced silence complex (RISC) to the 3-untranslated region (3-UTR) of mRNA targets [19]. Previous studies, including ours, have found that miR-34a-5p was obviously upregulated in NP tissues from IDD patients and it showed an ability to induce NP ECM degradation and cell apoptosis [17,20]. We also found that miR-34a-5p levels were dramatically upregulated in human NP cells under compression treatment in this study. Circular RNAs (circRNAs) are another class of endogenous ncRNAs that are rich in miRNA binding sites that act as competing endogenous RNAs (ceRNAs) and miRNA sponges, and thus regulate the target mRNAs translation and stability [21,22]. For instance, circSERPINE2 was reported to protect against osteoarthritis progression by functioning as a sponge for miR-1271, modulating the expression of E26 transformation-specific-related gene [23]. CircVMA21 was found to alleviate inflammatory cytokines-induced disc damage through miRNA-200c-XIAP pathway, as reported by Cheng et al. [24]. However, current knowledge about the biological roles of circRNAs regarding IVD integrity remains limited and vague. To the best of our knowledge, there is no research investigating whether circRNAs could modulate cell apoptosis and ECM degradation through interacting with miRNAs in human NP cells under mechanical stress.
In this study, we performed a circRNA microarray analysis in human NP cells without and with compression treatment and found that the circRNA involved in compression-induced damage of NP cells (circRNA-CIDN), which possessed a complementary sequence to miR-34a-5p seed region, was dramatically down-regulated under compression stress. Over-expression of circRNA-CIDN suppressed the apoptosis of NP cells and ECM degradation. Mechanistically, circRNA-CIDN bound to miR-34a-5p as a miRNA sponge to up-regulate the expression of silent mating type information regulation 2 homolog 1 (SIRT1). Furthermore, we adopted an ex vivo IVD organ culture model to reveal the role of circRNA-CIDN in mitigating compression-induced intervertebral disc degeneration. Our research demonstrated that circRNA-CIDN could be a promising therapeutic target for IDD.
2. Materials and methods
2.1. NP tissues collection
The control intervertebral disc tissues were collected from fifteen patients undergoing spinal surgery due to idiopathic scoliosis (IS). The degenerative intervertebral disc tissues were collected from thirty patients who underwent discectomy surgery for IDD. Detailed information for each patient was listed in Supplementary Table S1. The experimental protocols were approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology, and informed consent was obtained from each donor.
2.2. NP cells isolation and culture
The NP tissue samples were separated and the NP cells were isolated as described in previous work [25]. Briefly, NP tissues were cut into approximate 1 mm3 segments and digested enzymatically using 0.25% trypsin (Gibco, Life Technologies, Paisley, UK) for 0.5 h followed by 0.2% type II collagenase (Invitrogen, Carlsbad, CA, USA) for 3 h at 37 °C. After being filtered and washed using PBS, the suspension was centrifuged and the cells were cultured in Dulbecco's Modified Eagle's Medium with F12 nutrient mixture (Gibco, Grand Island, NY, USA), 15% foetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C in 5% CO2. When confluent, NP cells were digested by trypsinization and passaged for expansion, and cells from passage 2 were used in the subsequent experiments.
2.3. IVD organ model culture
The coccygeal discs (Co6/7, Co7/8) were harvested from male Sprague-Dawley rats (3 months old) provided by the Laboratory Animal Centre of Huazhong University of Science and Technology. Briefly, a total of twelve Sprague-Dawley rats with 24 total enrolled discs were euthanized using carbon dioxide, then their spinal columns were harvested en bloc under aseptic conditions. The soft tissues were removed and the enrolled coccygeal discs (Co6/7, Co7/8) were meticulously excised with intact endplates and the discs were flushed twice using PBS containing 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA). The study protocol was approved by the Animal Experimentation Committee of Huazhong University of Science and Technology. Entire discs with vertebral endplates were cultured in Dulbecco's Modified Eagle's Medium with F12 nutrient mixture, 15% FBS, and 1% penicillin/streptomycin. The culture medium was supplemented with 1.5% of a 0.4 M KCl and 5 M NaCl solution to keep the osmolarity at 400 mOsm/kg approximating physiological condition. The discs were incubated at 37 °C in 5% CO2. The medium was replaced twice a week.
2.4. Compression treatment
The human NP cells or ex vivo IVD organs were cultured in the compression apparatus with a stainless steel pressure vessel as described in our previous work [15]. The compression apparatus was used to deliver 1.0 MPa static compression. The pressure vessel was pumped into mixed 0.5% CO2 and 99.5% compressed air, and the cells or IVD organs were placed on six-well culture plates under humidified atmosphere at 37 °C. Unless otherwise specified in figures, the cells were under static compression for 36 h and the IVD organs were under compression treatment for 4 weeks.
2.5. RNA extraction, RT-PCR and qRT-PCR analysis
Total RNA was extracted from NP cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and obtained from enrolled tissue samples using TRIspin method in which the RNeasy column (Qiagen, Valencia, CA, USA) was used in the column purification steps as described previously [26]. RNA was reverse- transcribed using a Transcriptor First Strand cDNA Synthesis Kit (TAKARA Biotechnology, Otsu, Japan). The AmpliTaq DNA Polymerase (Life Technologies) was used for PCR. The cDNA and gDNA PCR products were run on 2% agarose gel electrophoresis. For cytoplasmic and nuclear RNA extracting, a Fisher BioReagents SurePrep Nuclear or Cytoplasmic RNA Purification Kit (Thermo Fisher, Waltham, MA, USA) was used according to merchant guide. The circRNA and mRNA levels were determined by quantitative PCR using SYBR Green assay; miRNAs levels were determined by mirVana qRT-PCR miRNA Detection Kit (Applied Biosystems, Foster City, USA), according to the manufacturer's protocols. The relative expression level of gene was calculated using 2—△△CT method. The mRNA and circRNA level were normalized to GAPDH, and miRNA level was normalized to U6. Primers used in this study are listed in supplementary Table S2.
2.6. RNase R treatment
Two micrograms of total RNA was incubated with or without RNase R (3 U/μg, epicenter) at 37 °C for 20 min. The resulting RNA was purified using a RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA).
2.7. CircRNA microarray analysis
Three samples were obtained from each of the two cell culture groups (control human NP cells vs. human NP cells with compression treatment for 36 h), then total RNA was extracted to perform microarray analysis. Detailed information of specimens used in circRNA microarray assay was indicated in Supplementary Table S1. The circRNAs microarray analysis was carried out as described previously [27]. The microarray data were analysed with GeneSpring GX software. We applied thresholds as follows: fold changes > 2, P values < 0.05 determined by t-test. To obtain the profiling of differentially expressed circRNAs between control and compression-treated human NP cells, the hierarchical clustering analysis were carried out according to the identified circRNAs levels by Cluster3.0 software. Differentially expressed circRNAs were shown by volcano plot filtering. The detailed information of the circRNA ID, host gene symbol, genome location and spliced sequence length were obtained from CircBase or DeepBase public database. The predicted functions of the differentially expressed circRNAs were analysed by Gene Ontology (GO) and Kyoto encyclopedia of Genes and Genomes (KEGG) pathway analysis of their host genes.
2.8. RNA immunoprecipitation (RIP)
The RIP experiment was performed with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA) with anti-AGO2 (#2897, Cell Signaling Technology, RRID:AB_2096291), following the manufacturer's instructions. IgG was used as a negative control. The AGO2 antibody was recovered with the protein A/G beads. Co-precipitated circRNA-CIDN and miR-34a-5p levels were evaluated by qRT–PCR analysis.
2.9. Dual luciferase assay
Human embryonic kidney (HEK) 293T cells were used for luciferase activity analysis and were plated on 96‐well plates, cultured to 60–70% confluence. For circRNA-CIDN and miR-34a-5p, the circRNA-CIDN fragments containing complementary binding sites for miR-34a-5p were inserted into a luciferase reporter vector. Wild-type pMIR-REPORT-circCIDN and mutant luciferase reporter pMIR-REPORT-circCIDN were synthesized by Obio Technology (Shanghai, China). 200 ng luciferase reporter vector of circCIDN-wt and circCIDN-mut, 50 nM miR-34a-5p and NC were transfected using lipofectamine 2000(Invitrogen). For SIRT1 and miR-34a-5p, the 3′UTR of human SIRT1 containing putative binding sites for miR-34a-5p were cloned into the vector. Wild-type pMIR-REPORT-SIRT1-3′UTR and mutant luciferase reporter pMIR-REPORT-SIRT1-3′UTR were synthesized by Obio Technology (Shanghai, China). 200 ng vector of SIRT1 3′UTR-wt and SIRT1 3′UTR -mut, 50 nM miR-34a-5p and NC were transfected. 48 h later, luciferase activity was examined with Dual Luciferase Reporter Assay kit (Promega) following the manufacturer's instructions.
2.10. Transfection vector construction and cell transfection
Recombinant adenovirus vector encoding circRNA-CIDN was synthesized by Obio Technology (Shanghai, China). The exons 1–5 of TRIM25 gene (1213 bp) with approximate 1 kb flanking intron sequences containing complementary Alu elements were amplified to construct circRNA-CIDN vector according to the method described previously with some modifications [28]. CircRNA-CIDN siRNA targeting the back-splice region and its negative control was synthesized by Obio Technology (Shanghai, China), and the target sequence for circRNA-CIDN was as follows: TGAAGAAGGTCTCCAGTTT. MiR-34a-5p mimics, miR-34a-5p inhibitors, and their corresponding negative controls were obtained from GenePharma (Shanghai, China). SIRT1 siRNA (5′-CCAUCUCUCUGUCACAAAUTT-3′) and scrambled siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from Ribo-bio (Guangzhou, China). When cultured NP cells grew to 80% confluence, transfections were performed using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocols. The transfections were performed up to 24 h before performing stimulations.
2.11. Intradiscal injection of circRNA-CIDN
Adenovirus circRNA-CIDN wt and mut were synthesized and packaged by Obio Technology, (Shanghai, China). Twenty-four ex vivo cultured IVD organs (from twelve male Sprague-Dawley rats, with two enrolled discs each) were randomly divided into four groups (control, compression treatment, circRNA-CIDN injection with compression, and circRNA-CIDN-mut injection with compression). 2 μl solution containing experimental or control virus vector (approximately 1 × 109 plaque-forming units per ml) overexpressing human circRNA-CIDN was slowly injected into the compression-treated discs with a 33-gauge needle at weeks 0 and 2. Except for control group, the IVD organs were under static compression treatment for four weeks.
2.12. Flow cytometry
Differently treated human NP cells were harvested and the apoptosis levels was determined using an Annexin V-APC/7-AAD Apoptosis Detection Kit (Yeasen Biotech, China) following manufacturer instructions. The NP cells were resuspended with binding buffer (5 × 105 cells per well) after washing by PBS, then incubated with Annexin V-APC and 7-AAD at room temperature for 15 min away from light. After labeling, samples were examined using a FACS Calibur flow cytometer (BD Biosciences, USA).
2.13. Western blotting analysis
After various treatments, the culture supernatants were removed and the cells were lysed using radio immunoprecipitation (RIPA) lysis buffer (Beyotime, China), then the concentrations of proteins were examined with an Enhanced BCA Protein Assay Kit (Beyotime, China). Equal amounts of protein from each sample were separated using 10–12% SDS-PAGE and transferred to PVDF membrane (Millipore, USA), which was blocked and incubated first with specific primary antibodies (1:500–1:1000) overnight at 4 °C and then with the appropriate horseradish peroxidase (HRP)-labeled secondary antibodies (1:2000; Abcam). Protein expression was observed with enhanced chemiluminescence reagents (Amersham, Piscataway, NJ, USA). Primary antibodies against these molecules were used: GAPDH (#5174, Cell signaling Technology, RRID:AB_10622025), SIRT1 (ab110304, Abcam, RRID:AB_10864359), Bax (ab32503, Abcam, RRID:AB_725631), Bcl-2 (ab196495, rabbit polyclonal antibody, Abcam, Cambridge, US), cleaved caspase-3 (#9664, Cell signaling Technology, RRID:AB_2070042), collagen II (ab34712, Abcam, RRID:AB_731688), aggrecan (13880-1-AP, Proteintech, RRID:AB_2722780), matrix metalloproteinase (MMP)−3 (#14351, Cell signaling Technology, RRID:AB_2798459), MMP-13 (ab39012, Abcam, RRID:AB_776416).
2.14. Histological assessment, immunohistochemistry, TUNEL staining and western blotting analysis in ex vivo IDD models
The rat IVDs were washed in PBS and harvested for further analyses. The specimens were fixed in formaldehyde, decalcified, dehydrated, embedded in paraffin, and sectioned in the midsagittal plane (perpendicular to endplates) at a thickness of 5 μm. Sections were stained with hematoxylin-eosin (HE) and safranin O-fast green (SO), and the histological scores were calculated based on the method previously described [29] to quantify the histological results. Immunohistochemistry was performed as described previously [25]. Briefly, sections were incubated with primary antibodies against SIRT1 (1:200), collagen II (1:200), MMP-13 (1:100), and cleaved caspase-3 (1:400) at 4 °C overnight, after which the sections were incubated with appropriate HRP-conjugated secondary antibodies and counterstained with hematoxylin. For terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) staining, the sections were handled using TUNEL Apoptosis Assay Kit (C1088, Beyotime). Nuclei were stained with DAPI (Beyotime) and samples were imaged with a fluorescence microscope (Olympus IX71, Tokyo, Japan). Western blotting in the NP tissues of IVDs samples was in the same steps as above.
2.15. Statistical analysis
Data are exhibited as means ± standard deviation of at least three independent experiments performed with samples from at least three different donors and data were processed using SPSS v.18.0 software (SPSS, Chicago, IL, USA). Differences between groups were evaluated using Student's t-test, Mann-Whitney U test or one-way analysis of variance (ANOVA) followed by the Tukey's test. A probability of less than 0.05 was considered statistically significant.
2.16. Data sharing
The raw data of circRNA microarray during the current study are available from the corresponding author on reasonable request.
3. Results
3.1. Identification of differentially expressed circRNAs in compression-treated human NP cells
To investigate the potential involvement of circular RNAs in compression-induced damage in human NP cells, we first examined the effects of compression treatment in cell samples from three different donors. As shown in Fig. S1a and b, compression treatment resulted in increased expression of MMP-3, MMP-13, Bax and cleaved caspase-3, decreased expression of collagen II, aggrecan and Bcl-2, and increased percentage of apoptotic cells. These results indicated the pro-apoptotic and pro-catabolic effect of compression in human NP cells, which is consistent with previous studies.
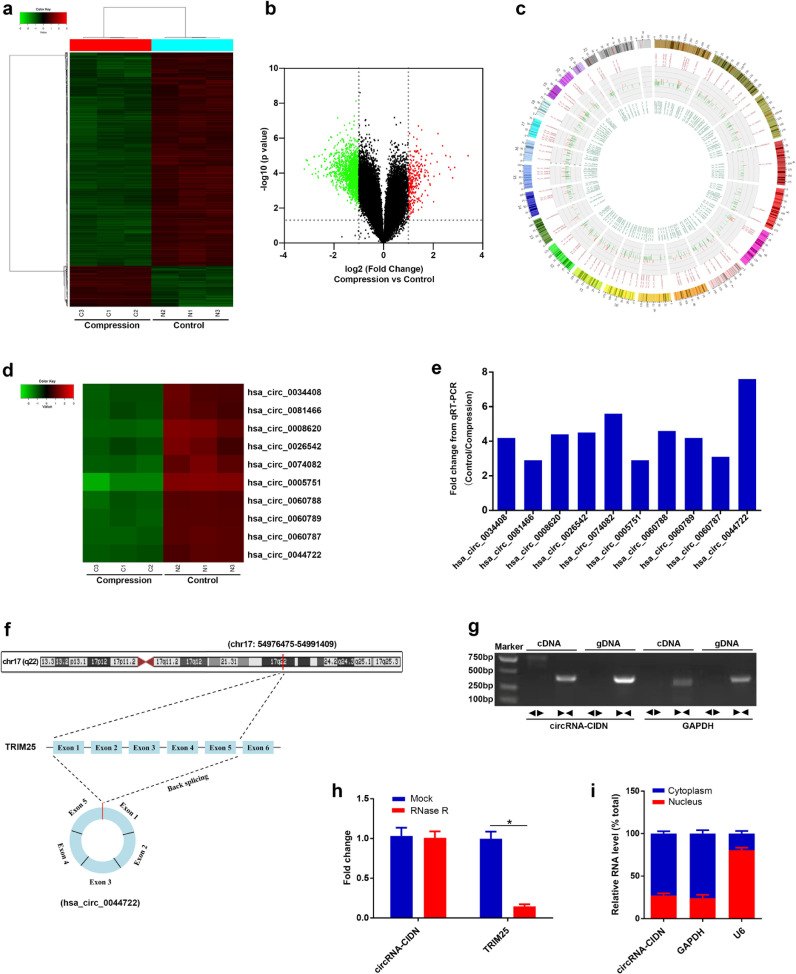
Next, we performed a circRNAs microarray analysis of human NP cells without and with compression treatment for 36 h. The results of the microarray analysis showed 1784 differentially expressed circRNAs between the two cell groups, of which 286 circRNAs were up-regulated and 1498 circRNAs were down-regulated in compression-treated human NP cells compared to controls, as represented on cluster heatmap (Fig. 1a). The variation of circRNAs expression was shown in volcano plots (Fig. 1b). The distribution of differentially expressed circRNAs on the human chromosomes was presented in a Circos plot, and we could see that the circRNAs were transcribed from all chromosomes (Fig. 1c). In addition, the potential functions of differentially expressed circRNAs were predicted by Gene Ontology (GO) and Kyoto encyclopedia of Genes and Genomes (KEGG) pathway analysis of their host genes. For the down-regulated circRNAs, the most significantly enriched and meaningful GO terms were related to regulation of transcription in biological process (BP), nucleoplasm part in cellular component (CC) and demethylase activity in molecular function (MF) (Fig. S1c). For the up-regulated circRNAs, the most significantly enriched GO terms were response to unfolded protein in BP, extracellular vesicle in CC and protein binding in MF (Fig. S1d). The top 30 most significantly enriched KEGG pathways associated with differentially expressed circRNAs were shown in Fig. S1e and f. Of these, the most significantly enriched and meaningful pathways were mTOR signaling pathway and JAK−STAT signaling pathway for the down-regulated circRNAs, and PI3K−Akt signaling pathway and AMPK signaling pathway for the up-regulated circRNAs.
Fig. 1.
CircRNA-CIDN was characterized as a compression related circular RNA in human NP cells. (a) Clustered heat map of the differentially expressed circRNAs in three samples from compression treated human NP cells and three control samples. Rows represented circRNAs identified by circRNA microarray, and columns represented cells specimens. The green indicates down-regulated circRNAs, and the red indicates up-regulated circRNAs. (b) The volcano plots showed the variation of circRNAs expression between two groups. Vertical lines expressed as 2.0-fold (log2 scaled) up or down changes; horizontal line represented the p value of 0.05 by Student's t-test (−log10 scaled). The red and green points in plot indicated the up-regulated and down-regulated circRNAs with statistical significance, respectively. (c) The distribution of differentially expressed circRNAs on the human chromosomes was presented on Circos plot. (d) Heat map analysis of ten significantly down-regulated circRNAs identified in this study. (e) The fold-changes of the ten significantly down-regulated circRNAs obtained from the qRT-PCR results. Cell samples from three different donors were tested (n = 3). All the three samples were different from that for the microarray analysis. (f) Schematic of the genomic loci of TRIM25 gene and circRNA-CIDN. The genomic coordinates referred to the human genome reference hg19/GRCh37. (g) The products amplified using divergent (←→) or convergent (→←) primers were verified by agarose gel electrophoresis. Divergent-circRNA-CIDN primers amplified circRNA-CIDN in cDNA but not gDNA. Convergent-circRNA-CIDN primers amplified circRNA-CIDN in both cDNA and gDNA. GAPDH could only be amplified using Convergent-GAPDH primers, in both cDNA and gDNA. The primers information was given in supplementary Table S2. (h) Relative abundance of circRNA-CIDN and TRIM25 mRNA in human NP cells with or without RNase R treatment were examined with qRT-PCR analysis. *p < 0.05 versus mock treatment for TRIM25 group, n = 3 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD). (i) qRT-PCR assay in nuclear and cytoplasmic fractions showed the levels of circRNA-CIDN, cytoplasmic control transcript (GAPDH mRNA) and nuclear control transcript (U6).
3.2. Hsa_circ_0044722 (circRNA-CIDN) was identified as a compression related circRNA in human NP cells
Considering the up-regulated circRNAs were the minority of the differentially expressed circRNAs, we then confirmed the microarray analysis results of ten significantly down-regulated circRNAs in compression-treated human NP cells and control groups using qRT-PCR. As shown in Fig. 1d, the heat map analysis revealed these ten circRNAs expression patterns according to the circRNA microarray. The log2 fold-changes of these circRNAs from the qRT-PCR validation results were calculated (Fig. 1e). The expression patterns of these ten circRNAs from qRT-PCR results were consistent with the microarray analysis. Because the qRT-PCR results showed that hsa_circ_0044722 is the most down-regulated circRNAs with a greater than 7-fold change, we chose hsa_circ_0044722 for further research. According to the CircBase database annotation, hsa_circ_0044722 is derived from the exons 1 to 5 of TRIM25 gene (chr17: 54976475–54991409) (Fig. 1f). The length of hsa_circ_0044722 is 1213 bp. We termed this circRNA involved in compression-induced damage of NP cells as “circRNA-CIDN”. Using cDNA and genomic DNA (gDNA) from human NP cells as templates, we validated that circRNA-CIDN could only be amplified by divergent primers in cDNA, and no products were observed in the gDNA groups (Fig. 1g). Additionally, RNase R exonuclease was used to further validate circRNA-CIDN in human NP cells. Resistance to digestion by RNase R exonuclease confirmed that this RNA species was indeed circular (Fig. 1h). Moreover, the RT-qPCR assay in nuclear and cytoplasmic fractions showed a predominant cytoplasmic distribution of circRNA-CIDN in human NP cells (Fig. 1i).
3.3. CircRNA-CIDN inhibited compression-induced apoptosis and ECM degradation of human NP cells
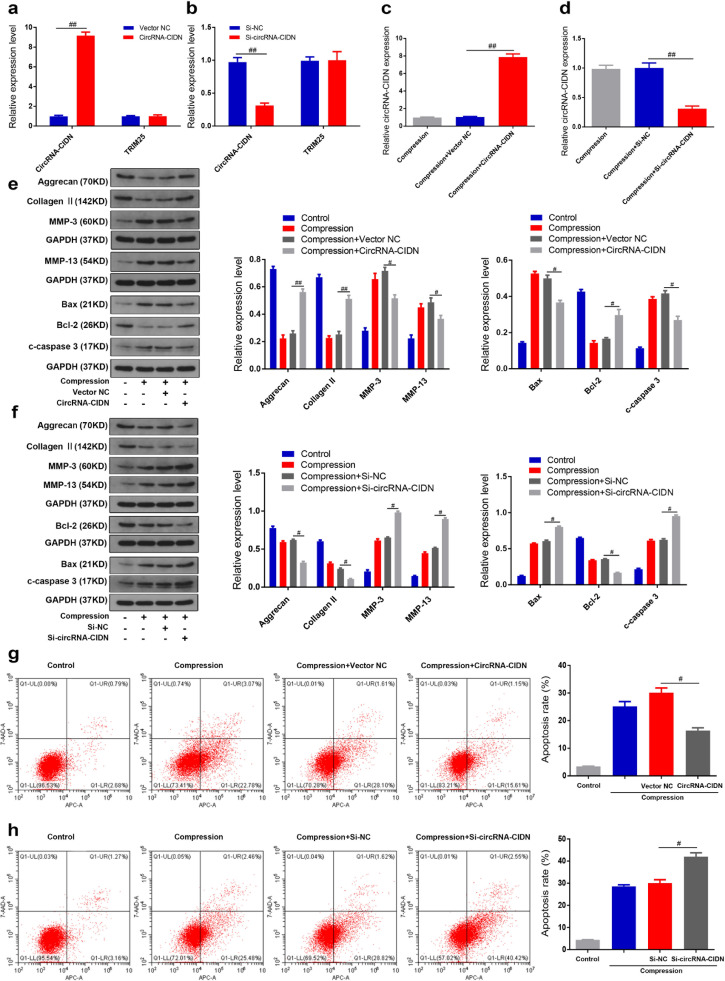
To explore the functions of circRNA-CIDN in compression-treated human NP cells, we established a circRNA-CIDN expression adenovirus vector to generate NP cells that stably overexpressed circRNA-CIDN. As shown in Fig. 2a, the expression of circRNA-CIDN was significantly increased in human NP cells transfected with circRNA-CIDN adenovirus vector. In contrast, when we transfected circRNA-CIDN siRNA targeting the junction site of circRNA-CIDN into human NP cells, the qRT-PCR results showed that circRNA-CIDN siRNA inhibited the expression of circRNA-CIDN, but had no effect on the expression of the TRIM25 mRNA level (Fig. 2b). Under compression treatment, NP cells transfected with circRNA-CIDN expression adenovirus vector or circRNA-CIDN siRNA showed higher and lower circRNA-CIDN expression levels, respectively, when compared with NP cells treated with compression alone (Fig. 2c and d). Next, the western blot analysis revealed that circRNA-CIDN knockdown significantly increased the up-regulation of MMP-3, MMP-13, Bax, cleaved caspase-3 expression and the down-regulation of collagen II, aggrecan, Bcl-2 expression induced by compression. CircRNA-CIDN overexpression partially reversed the effects of compression on these gene expression (Fig. 2e and f). The effect of circRNA-CIDN on apoptosis of compression-treated human NP cells was further examined by flow cytometry analysis with Annexin V/PI dual staining. The results of flow cytometry indicated that circRNA-CIDN overexpression obviously suppressed compression-induced apoptosis of NP cells (Fig. 2g). Conversely, circRNA-CIDN inhibition remarkably enhanced the apoptotic effects of compression (Fig. 2h). Taken together, our data suggest that circRNA-CIDN could have a protective role in human NP cells exposed to compression.
Fig. 2.
CircRNA-CIDN inhibited compression-induced NP cell apoptosis and ECM degradation. (a,b) The RNA levels of circRNA-CIDN and TRIM25 in human NP cells transfected with circRNA-CIDN adenovirus or circRNA-CIDN siRNA were measured by qRT-PCR. ##p < 0.01 versus NC group, n = 3 [Student's t-test]. (c,d) Human NP cells under compression treatment were transfected with circRNA-CIDN adenovirus or circRNA-CIDN siRNA, and the expression levels of circRNA-CIDN were measured by qRT-PCR. ##p < 0.01 versus compression + NC group, n = 3 [Student's t-test]. (e-f) Human NP cells under compression were transfected with circRNA-CIDN adenovirus or circRNA-CIDN siRNA, and the protein levels of aggrecan, collagen II, MMP-3, MMP-13, Bax, Bcl-2 and c-caspase 3 were measured by western blotting. The data were normalized to GAPDH expression levels. #p < 0.05, ##p < 0.01 versus compression + NC group, n = 3 [Student's t-test]. (g-h) Human NP cells under compression were transfected with circRNA-CIDN adenovirus or circRNA-CIDN siRNA, and the rate of apoptosis was detected by Annexin V-APC/7-AAD staining. #p < 0.05 versus compression + NC group, n = 3 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD).
3.4. CircRNA-CIDN served as a sponge for miR-34a-5p in human NP cells
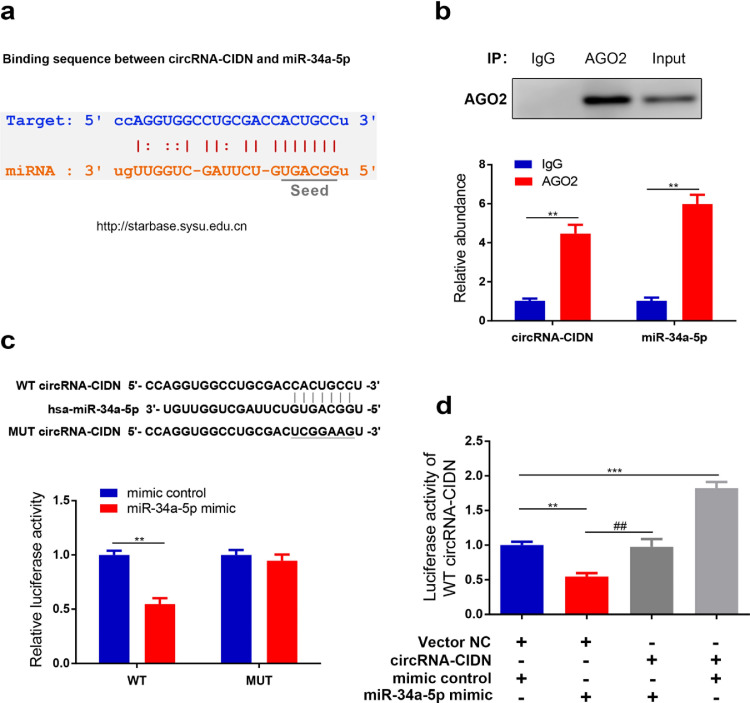
Given that circRNAs have been shown to act as miRNAs sponges and that circRNA-CIDN is a stable molecule and primarily located in the cytoplasm, we speculated that circRNA-CIDN could also target a specific miRNA and modulate its downstream functions. According to the StarBase online database, circRNA-CIDN possesses a complementary sequence to miR-34a-5p seed region (Fig. 3a), therefore we conducted the following experiments to verify whether circRNA-CIDN acts as a sponge of miR-34a-5p in human NP cells. Firstly, since it is well known that miRNAs degrade mRNA and inhibit translation in an AGO2-dependent manner via directly binding to their targets, we thus performed an anti-AGO2 RIP in human NP cells. The results showed that circRNA-CIDN and miR-34a-5p pulled down by anti-AGO2 antibodies were both significantly enriched, indicating that circRNA-CIDN and miR-34a-5p existed in RISC (Fig. 3b). Secondly, a circRNA-CIDN fragment with wild type (WT) or mutant (MUT) complementary binding sites for miR-34a-5p was constructed and inserted into pMIR-REPORT luciferase reporter vectors. The results of luciferase reporter assay showed that miR-34a-5p overexpression significantly suppressed the luciferase activity of WT reporter but not the MUT reporter, suggesting that circRNA-CIDN was directly bound by miR-34a-5p via complementary target sites (Fig. 3c). Moreover, the inhibition of the luciferase activity of WT reporter induced by miR-34a-5p was rescued by circRNA-CIDN overexpression (Fig. 3d). Overall, these findings suggested that circRNA-CIDN may function as a miRNAs sponge for miR-34a-5p in human NP cells.
Fig. 3.
CircRNA-CIDN acted as a sponge of miR-34a-5p. (a) Schematic of the predicted miR-34a-5p site in the circRNA-CIDN from StarBase online database. (b) The amount of circRNA-CIDN and miR-34a-5p in human NP cells were examined by RIP assay. AGO2 was determined by IP-western blotting (up); circRNA-CIDN and miR-34a-5p expression levels were determined by qRT-PCR (down). ⁎⁎p < 0.01 versus control group, n = 3 [Student's t-test]. (c) Luciferase reporter activity of circRNA-CIDN in HEK-293T cells co-transfected with miR-34a-5p mimic or mimic control. ⁎⁎p < 0.01 versus mimic control group, n = 3 [Student's t-test]. (d) Luciferase reporter activity of WT circRNA-CIDN was analysed after co-transfection with circRNA-CIDN or Vector NC, miR-34a-5p mimic or mimic control. ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001 versus Vector NC + mimic control group, ##p < 0.01 versus Vector NC + miR-34a-5p mimic group, n = 3 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD).
3.5. MiR-34a-5p enhanced compression-induced apoptosis and ECM degradation of human NP cells
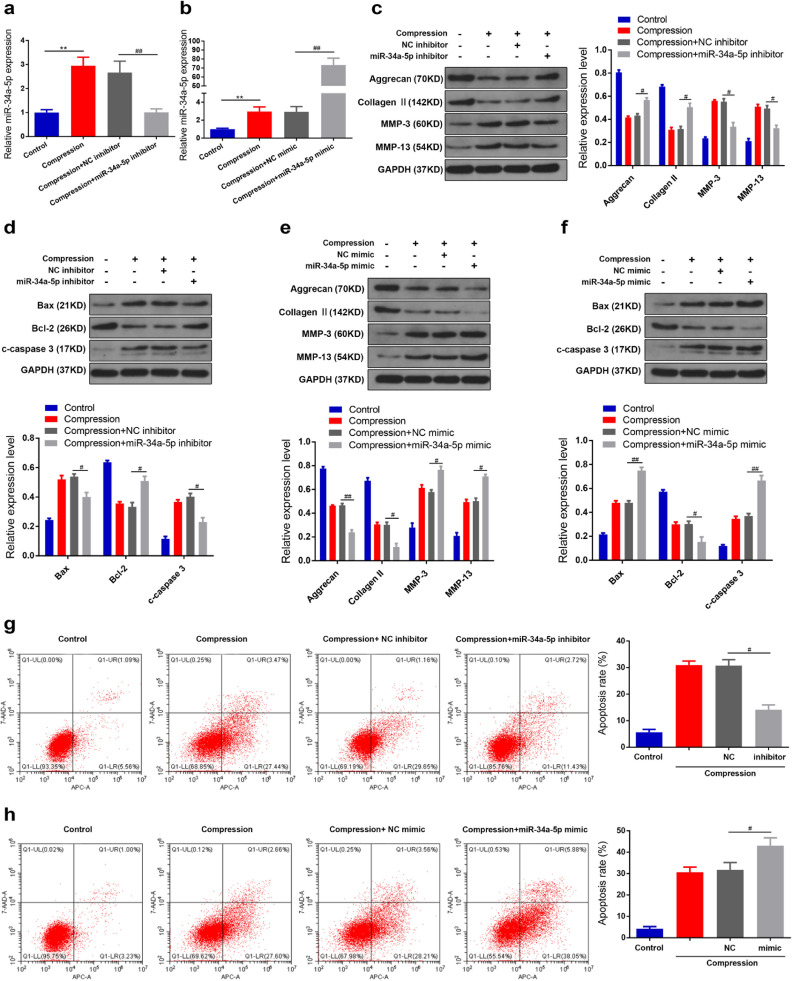
Considering the present findings that miR-34a-5p is the circRNA-CIDN-associated miRNA in human NP cells, we hypothesized that miR-34a-5p might also exert an important role in compression-stimulated human NP cells. The qRT-PCR results showed that compression treatment increased miR-34a-5p levels in human NP cells, which could be suppressed and promoted by miR-34a-5p inhibitor and miR-34a-5p mimic, respectively (Fig. 4a and b). Functionally, the enhanced apoptosis and changes in ECM metabolism in response to compression were markedly suppressed after miR-34a-5p knockdown. In contrast, under compression stimulation, apoptosis and ECM metabolism changes increased when the cells were transfected with a miR-34a-5p mimic (Fig. 4c–h).
Fig. 4.
MiR-34a-5p enhanced compression-induced NP cell apoptosis and ECM degradation. (a,b) Human NP cells under compression were transfected with miR-34a-5p inhibitor or miR-34a-5p mimic, and RNA levels of miR-34a-5p were measured by qRT-PCR. ⁎⁎p < 0.01 versus control group, ##p < 0.01 versus compression + NC group, n = 3 [Student's t-test]. (c,d) The protein levels of aggrecan, collagen II, MMP-3, MMP-13, Bax, Bcl-2 and c-caspase 3 in human NP cells transfected with miR-34a-5p inhibitor were measured by western blotting. Proteins were normalized to GAPDH expression levels. #p < 0.05 versus compression + NC group, n = 3 [Student's t-test]. (e-f) The protein levels in human NP cells transfected with miR-34a-5p mimic were measured by western blotting. #p < 0.05, ##p < 0.01 versus compression + NC group, n = 3 [Student's t-test]. (g-h) Human NP cells under compression were transfected with miR-34a-5p inhibitor or miR-34a-5p mimic, and the apoptosis rate was detected by Annexin V-APC/7-AAD staining. #p < 0.05 versus compression + NC group, n = 3 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD).
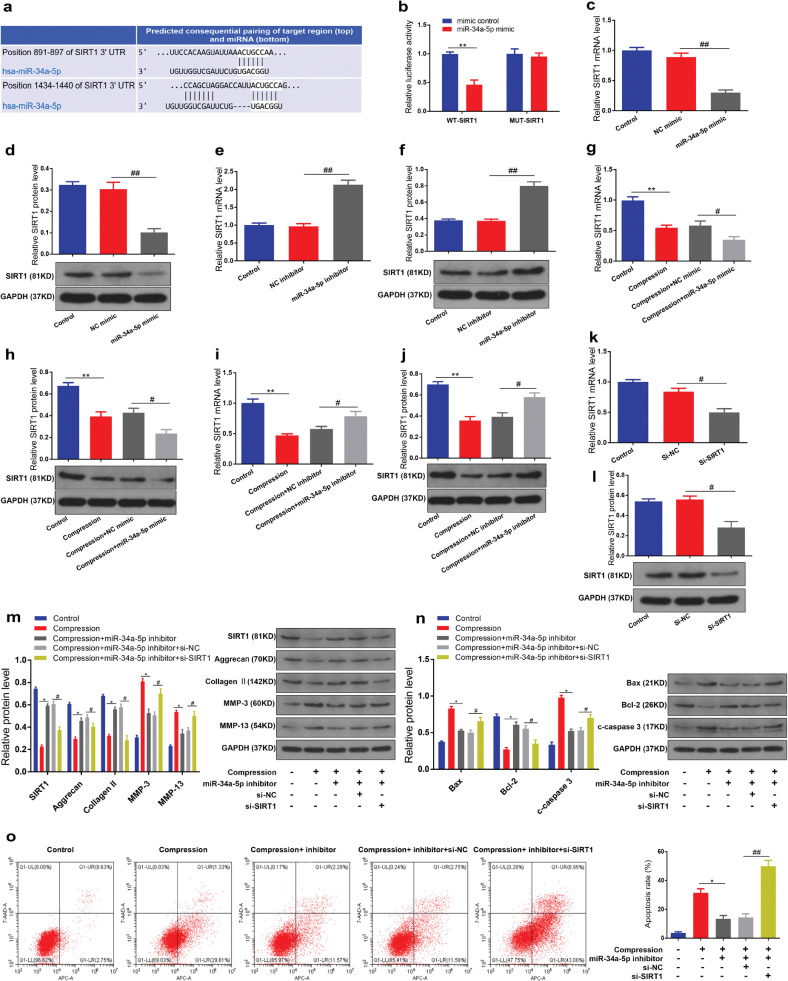
3.6. MiR-34a-5p repressed SIRT1 expression by targeting 3′-UTR of SIRT1
Using the TargetScan online prediction database, we found that silent mating type information regulation 2 homolog 1 (SIRT1), a critical regulator of cell differentiation, proliferation, and apoptosis, is a potential target of miR-34a-5p (Fig. 5a). To confirm this finding, we constructed a luciferase reporter vector with the wild type (WT) or mutant (MUT) SIRT1 3′-UTR possessing the putative miR-34a-5p target site. The data showed that miR-34a-5p overexpression obviously inhibited the luciferase activity of the reporter containing the WT 3′-UTR of SIRT1 compared to mimic control, whereas no significant change in the luciferase activity was detected in the MUT 3′-UTR of SIRT1 group (Fig. 5b). Moreover, qRT-PCR and western blot assay were performed to further confirm the luciferase reporter gene assay results. We found that miR-34a-5p overexpression remarkably inhibited SIRT1 mRNA and protein expression in human NP cells (Fig. 5c and d). In contrast, downregulation of miR-34a-5p noticeably increased SIRT1 expression level (Fig. 5e and f). These data indicated that miR-34a-5p could directly bind to the 3′-UTR of SIRT1 and negatively regulate its expression. Subsequently, we considered whether SIRT1 was involved in the effects of miR-34a-5p on compression-stimulated human NP cells. The qRT-PCR and western blot assay were performed to investigate the effect of compression on SIRT1 expression in human NP cells, and the data indicated that compression treatment resulted in a decrease of SIRT1 mRNA and protein expression (Fig. 5g–j). In addition, miR-34a-5p overexpression facilitated the suppressive effect of compression treatment on SIRT1 expression while miR-34a-5p inhibition partially attenuated the effect of compression treatment on SIRT1 expression (Fig. 5g–j). Furthermore, we use siRNA to knock down SIRT1 in human NP cells (Fig. 5k and l). The absence of SIRT1 markedly counteracted the effects of miR-34a-5p inhibition in human NP cells treated by compression (Fig. 5m–o). Collectively, these results suggested that miR-34a-5p exerted its functions by targeting SIRT1.
Fig. 5.
SIRT1 was a direct target gene of miR-34a-5p. (a) Schematic of the predicted miR-34a-5p binding sites in the 3′UTR of SIRT1 mRNA from TargetScan online database. (b) Luciferase reporter activity of SIRT1 in HEK-293T cells co-transfected with miR-34a-5p mimic or mimic control. **p < 0.01 versus mimic control group, n = 3 [Student's t-test]. (c) SIRT1 mRNA level in human NP cells transfected with miR-34a-5p mimic was measured by qRT-PCR. ##p < 0.01 versus NC group, n = 3 [Student's t-test]. (d) SIRT1 protein level in human NP cells transfected with miR-34a-5p mimic was measured by western blotting. Proteins were normalized to GAPDH expression levels. ##p < 0.01 versus NC group, n = 3 [Student's t-test]. (e,f) SIRT1 mRNA and protein level in human NP cells transfected with miR-34a-5p inhibitor. ##p < 0.01 versus NC group, n = 3 [Student's t-test]. (g,h) SIRT1 mRNA and protein level in human NP cells transfected with miR-34a-5p mimic under compression treatment. **p < 0.01 versus control group, #p < 0.05 versus compression + NC group, n = 3 [Student's t-test]. (i,j) SIRT1 mRNA and protein level in human NP cells transfected with miR-34a-5p inhibitor under compression treatment. **p < 0.01 versus control group, #p < 0.05 versus compression + NC group, n = 3 [Student's t-test]. (k,l) SIRT1 mRNA and protein level in human NP cells transfected with SIRT1 siRNA. #p < 0.05 versus NC group, n = 3 [Student's t-test]. (m,n) The protein levels of SIRT1, aggrecan, collagen II, MMP-3, MMP-13, Bax, Bcl-2 and c-caspase 3 were measured by western blotting. *p < 0.05 versus compression group, #p < 0.05 versus compression + miR-34a-5p inhibitor + si-NC group, n = 3 [Student's t-test]. (o) The apoptosis rate was detected by Annexin V-APC/7-AAD staining. *p < 0.05 versus compression group, ##p < 0.01 versus compression + miR-34a-5p inhibitor + si-NC group, n = 3 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD).
3.7. CircRNA-CIDN modulated compression-induced apoptosis and ECM degradation of human NP cells by the miR-34a-5p/SIRT1 pathway
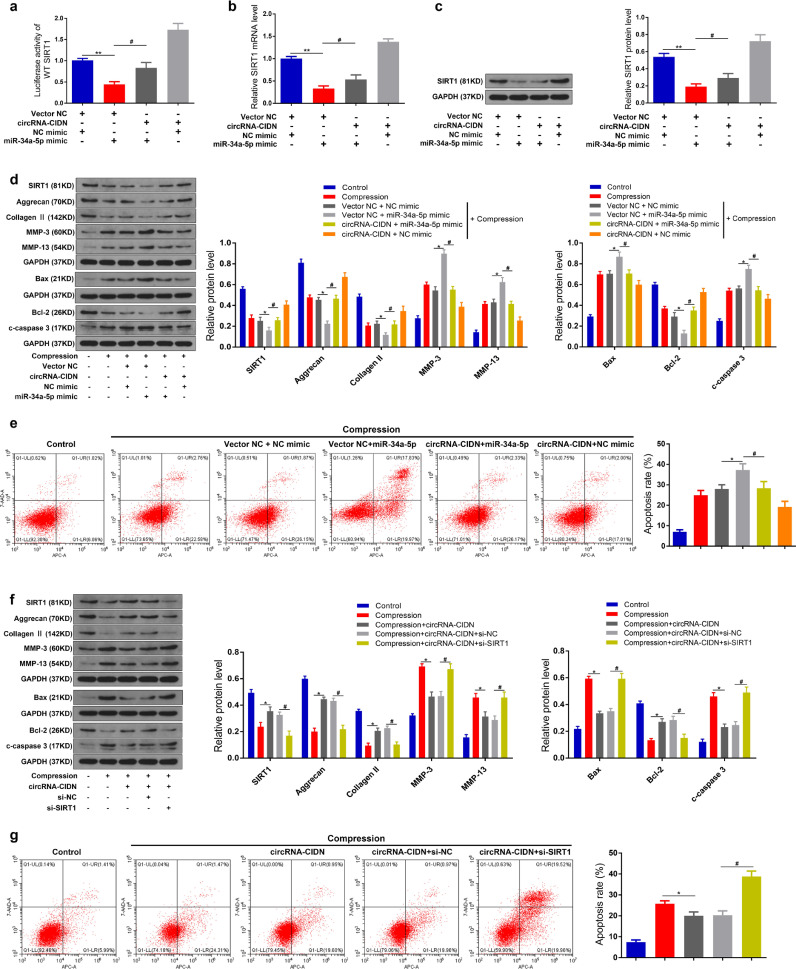
After confirming the relationships between circRNA-CIDN and miR-34a-5p, and miR-34a-5p and SIRT1, we further explored the combined effect of circRNA-CIDN and miR-34a-5p on compression-induced apoptosis and ECM degradation of human NP cells, as well as the involvement of SIRT1. We first performed a luciferase reporter assay, qRT-PCR and western blot to detect the effect of circRNA-CIDN on miR-34a-5p-mediated repression of the SIRT1 gene. The results showed that the luciferase activity of WT 3′-UTR of SIRT1 reporter was significantly decreased in the miR-34a-5p mimic group compared with the control group. However, co-transfection with miR-34a-5p mimic and circRNA-CIDN overexpression adenovirus vector was able to rescue this suppressive effect (Fig. 6a). Consistently, the inhibition in SIRT1 mRNA and protein expression caused by miR-34a-5p overexpression could also be abolished by circRNA-CIDN overexpression in human NP cells (Fig. 6b and c). These results indicated that circRNA-CIDN overexpression neutralized miR-34a-5p-mediated SIRT1 repression in human NP cells.
Fig. 6.
CircRNA-CIDN functioned in compression treated NP cells by targeting miR-34a-5p and SIRT1. (a) Luciferase reporter activity of WT SIRT1 was analysed after co-transfection with circRNA-CIDN or Vector NC, miR-34a-5p mimic or NC mimic. ⁎⁎p < 0.01 versus Vector NC + NC mimic group, #p < 0.05 versus Vector NC + miR-34a-5p mimic group, n = 3 [Student's t-test]. (b) SIRT1 mRNA level in human NP cells under compression was measured by qRT-PCR. ⁎⁎p < 0.01 versus Vector NC + NC mimic group, #p < 0.05 versus Vector NC + miR-34a-5p mimic group, n = 3 [Student's t-test]. (c) The protein level of SIRT1 in human NP cells under compression was measured by western blotting. ⁎⁎p < 0.01 versus Vector NC + NC mimic group, #p < 0.05 versus Vector NC + miR-34a-5p mimic group, n = 3 [Student's t-test]. (d) The protein levels of SIRT1, aggrecan, collagen II, MMP-3, MMP-13, Bax, Bcl-2 and c-caspase 3 were measured by western blotting. Proteins were normalized to GAPDH expression levels. *p < 0.05 versus Vector NC + NC mimic group, #p < 0.05 versus Vector NC + miR-34a-5p mimic group, n = 3 [Student's t-test]. (e) The apoptosis rate was detected by Annexin V-APC/7-AAD staining. *p < 0.05 versus Vector NC + NC mimic group, #p < 0.05 versus Vector NC + miR-34a-5p mimic group, n = 3 [Student's t-test]. (f) The protein levels of SIRT1, aggrecan, collagen II, MMP-3, MMP-13, Bax, Bcl-2 and c-caspase 3 were measured by western blotting. *p < 0.05 versus compression group, #p < 0.05 versus compression + circRNA-CIDN + si-NC group, n = 3 [Student's t-test]. (g) The apoptosis rate was detected by Annexin V-APC/7-AAD staining. *p < 0.05 versus compression group, #p < 0.05 versus compression + circRNA-CIDN + si-NC group, n = 3 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD).
Subsequently, we conducted western blot and flow cytometry analysis with Annexin V/PI dual staining. The result indicated that upregulation of miR-34a-5p facilitated the pro-apoptotic and pro-catabolic effect of compression treatment, and such facilitative effect of miR-34a-5p overexpression was blocked by circRNA-CIDN overexpression (Fig. 6d and e). Additionally, circRNA-CIDN expression adenovirus vector and siRNA-mediated SIRT1 silencing (si-SIRT1) were co-transfected to examined whether SIRT1 was the downstream mediator of circRNA-CIDN. In compression-treated human NP cells, upregulation of circRNA-CIDN prevented cell apoptosis and ECM degradation, while knockdown of SIRT1 impaired this protective effect (Fig. 6f and g). Taken together, these results indicated that circRNA-CIDN overexpression could protect human NP cells from the attack of compression treatment by regulating miR-34a-5p and SIRT1.
3.8. Validation of the expression of circRNA-CIDN, miR-34a-5p and SIRT1 mRNA in human degenerative and healthy NP tissues
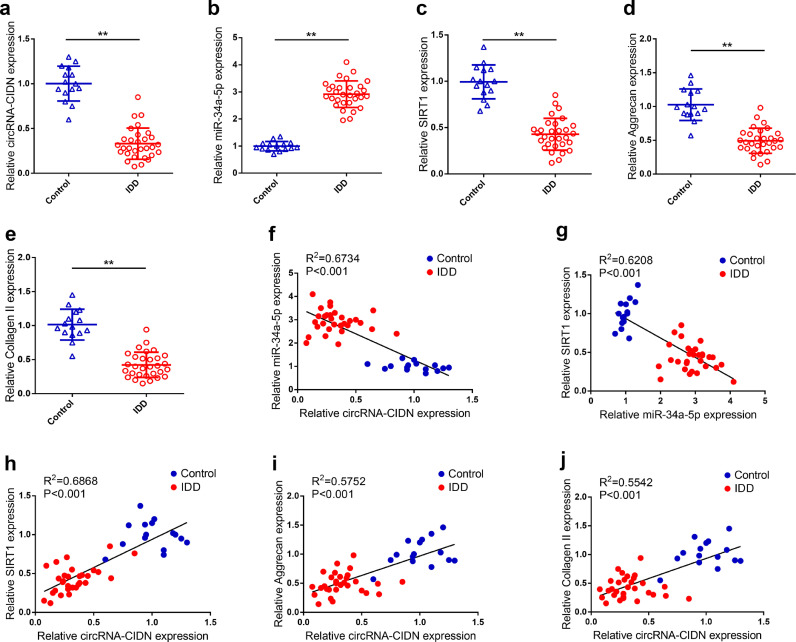
To further confirm the above results, we validated the expression levels of circRNA-CIDN, miR-34a-5p and SIRT1 genes in human NP tissues by qRT-PCR. As shown in Fig. 7a–c, circRNA-CIDN and SIRT1 mRNA expression was down-regulated while miR-34a-5p expression was up-regulated in degenerative NP tissues compared with that in control NP tissues. Furthermore, there was a negative correlation between circRNA-CIDN and miR-34a-5p levels, suggesting that the enhanced destructive effects of miR-34a-5p in compression-stimulated human NP cells may be due to the down-regulation of circRNA-CIDN (Fig. 7f). We also found that miR-34a-5p expression was negatively correlated with SIRT1 mRNA expression in NP tissues, indicating that miR-34a-5p could directly inhibit SIRT1 mRNA expression (Fig. 7g). In addition, we observed a positive correlation between circRNA-CIDN expression and SIRT1 mRNA expression, demonstrating that up-regulation of circRNA-CIDN could abrogate the repressive effect of miR-34a-5p on its downstream target SIRT1 gene (Fig. 7h). It is worth noting that we also monitored the expression levels of aggrecan and collagen II, and found that aggrecan and collagen II mRNA expressions were significantly down-regulated in degenerative NP tissues compared to that in control NP tissues (Fig. 7d and e). The circRNA-CIDN levels, and the aggrecan and collagen II mRNA expression levels demonstrated a significant positive correlation (Fig. 7i and j). Taken together, these results supported the notion that circRNA-CIDN might function as a novel protective factor in compression-stimulated human NP cells through the miR-34a-5p/SIRT1 axis.
Fig. 7.
Validation of the expression of circRNA-CIDN, miR-34a-5p and SIRT1 in human IDD tissue specimens. (a–e) The expression levels of circRNA-CIDN, miR-34a-5p, SIRT1, aggrecan and collagen II mRNA in 15 control (idiopathic scoliosis) and 30 IDD tissue specimens was measured by qRT-PCR. ⁎⁎p < 0.01 [Student's t-test]. Data presented as means with error bars representing standard deviation (SD). (f–j) The correlation between circRNA-CIDN and miR-34a-5p, SIRT1, aggrecan and collagen II, correlation between miR-34a-5p and SIRT1 mRNA level were analysed by Spearman's rank correlation analysis.
3.9. CircRNA-CIDN ameliorated IDD development in an ex vivo IDD model
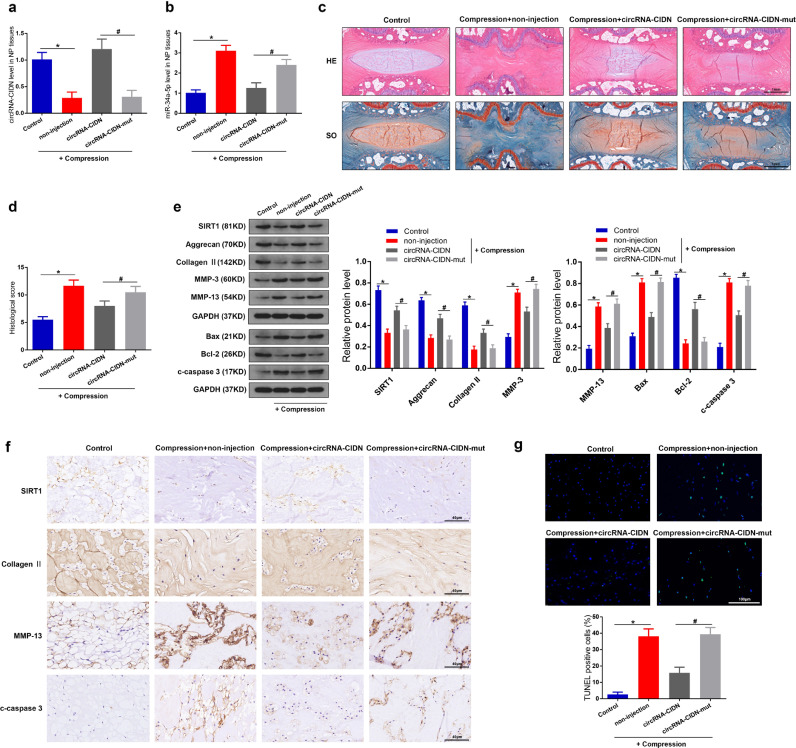
Considering all the experimental results above were obtained in vitro, we further applied an ex vivo IVD organ culture model to validate the role of circRNA-CIDN in compression induced IDD. Adenovirus circRNA-CIDN wt or mut was injected into the ex vivo rat IVDs, which were cultured in a compression apparatus to induce IDD progression. The efficiency of circRNA-CIDN overexpression was confirmed by RT-qPCR analysis (Fig. 8a) and the miR-34a-5p level in the degenerative NP tissues was remarkably downregulated after circRNA-CIDN overexpression (Fig. 8b). Then we assessed the histomorphological changes in the IVD tissues by using HE and SO staining (Fig. 8c): the oval-shaped NP constituted a large volume of the disc in the midsagittal cross-section and contained a large amount of glycosaminoglycan in control group, while the IDD group showed a collapse of the disc height together with notable fibrous-tissue invasion. The injection of adenoviral circRNA-CIDN alleviated the loss of NP tissue and the destruction of disc structure. The histological score of the adenoviral circRNA-CIDN group was obviously lower than that of the IDD group (Fig. 8d). Moreover, the overexpression of circRNA-CIDN downregulated the expression of ECM catabolism enzymes (MMP-3, MMP-13) and pro-apoptotic genes (Bax and cleaved caspase-3), and upregulated the level of anabolism markers (collagen II, aggrecan) and anti-apoptotic protein (Bcl-2) in the ex vivo compression-induced IDD model (Fig. 8e). Immunohistochemical analysis and apoptosis detection by TUNEL staining of the IVDs further validated that circRNA-CIDN injection could mitigate the degenerative changes of the NP in the IDD model (Fig. 8f and g). Together, these findings demonstrated that circRNA-CIDN retarded IDD progression ex vivo.
Fig. 8.
CircRNA-CIDN retarded compression-induced IDD ex vivo. (a) qRT-PCR showed that the downregulated circRNA-CIDN level in the compression treated IVDs was rescued by the injection of circRNA-CIDN. (b) The RNA level of miR-34a-5p was measured by qRT-PCR. (c) Hematoxylin-eosin (HE) and Safranin O-fast green (SO) staining of the IVDs samples. Scale bar: 1 mm. (d) The histological scores of the IVDs according to histological grading scale. (e) The protein levels of SIRT1, aggrecan, collagen II, MMP-3, MMP-13, Bax, Bcl-2 and c-caspase 3 in the NP tissues of IVDs were measured by western blotting. Proteins were normalized to GAPDH expression levels. (f) Immunohistochemical analysis showed the expression of SIRT1, collagen II, MMP-13 and cleaved-caspase3 proteins in the NP tissues. Scale bar: 40 μm. (g) TUNEL staining and fluorescence microscope analysis were used to detected apoptosis in the IVDs samples. The green fluorescence indicated TUNEL positive cells. Scale bar:100 µm. *p < 0.05 versus control group, #p < 0.05 versus circRNA-CIDN-mut group, n = 6 [Mann–Whitney U test]. Data presented as means with error bars representing standard errors of the mean (SEM).
4. Discussion
To date, therapeutic alternatives for IDD are limited and the clinical treatment efficacy is far from satisfactory, which is highly related to the fact that the pathogenic mechanisms of IDD remain unclear. Thus, further exploration of the pathogenesis underlying IDD is crucial to develop accurate pathophysiological diagnosis and elaborate precise therapeutic regimens [4]. The IVD is subjected to numerous mechanical loads in vivo, which plays an important role in the process of IDD development [14]. Abnormal mechanical loads have been demonstrated to induce various pathological alterations within the IVD, including the two major causes of IDD, namely NP cell apoptosis and ECM destruction, impairing the physiological functions of IVD [15,30,31]. In this study, we observed increased expression of ECM catabolism enzymes (MMP-3, MMP-13), pro-apoptotic genes (Bax and cleaved caspase-3), decreased expression of ECM anabolism markers (collagen II, aggrecan) and anti-apoptotic protein Bcl-2, and increased percentage of apoptotic cells in the compression-induced NP cell model. The cells exposed to compression loading could not be exactly the same as the cells from in vivo IDD conditions, in which the microenvironment was more complex with oxidative stress and local inflammation, et cetera [25]. However, the above results indicated that treating mechanical stress on disc cells effectively simulated the pathological responses during IDD process. In the present study, the NP cells under mechanical loading served a valuable experimental platform for researching the involvement of compression in IDD pathophysiology. Moreover, studies have reported that a static or dynamic loading organ culture could effectively simulate the cellular response to mechanical stress within IVD [32], [33], [34]. Compared to in vitro assays, IVD whole organ culture offered various improvements in deciphering the pathogenic mechanisms underlying IDD, including the intactness of IVD structure with native ECM which was crucial to demonstrate compression-induced IVD degeneration [35], [36], [37]. For this reason, we also adopted an ex vivo IVD organ culture model to mimic the in vivo IVD environment for further research.
Among the non-coding RNA superfamily members found to be involved in the initiation and progression of IDD, circRNA is a novel class of endogenous RNAs that has a unique structure of covalently closed loop, as well as cell type-specific and tissue-specific expression pattern [38]. While accumulating studies have reported that circRNAs may also be central regulators of biological and pathological processes [39], [40], [41], its potential role in IDD still needs further research. There were some evidence suggesting that the expression of circRNAs was dysregulated in IDD tissues and several specific circRNAs were found to functionally participate in the initiation and development of degenerative disc diseases [42]. For example, circRNA_104670 was upregulated in degenerative NP tissues and contributed to IDD progress through sponging miR-17–3p to impair NP cell survival and promote ECM degradation [43]. CircVMA21 and circSEMA4B were found to be downregulated in IDD tissue specimens and they could protect the IVD through competitive inhibition of microRNA [24,44]. However, the previous studies about how circRNAs functioned in the pathogenesis of IDD did not shed light on the involvement of mechanical stress. To systematically investigate the role of circRNAs in compression loads induced IDD, we performed circRNAs microarray analysis in human NP cells without and with static compression. To the best of our knowledge, this was the first study on the circRNAs expression profile in compression loads induced IDD. We identified 286 up-regulated circRNAs and 1498 down-regulated circRNAs in compression treatment groups compared to control groups. Then the key down-regulated circRNA, circRNA-CIDN was characterized, which was further validated by the qRT-PCR assay showing a greater than seven-fold down-regulation change. Subsequently, gain-of-function and loss-of-function assays revealed circRNA-CIDN obviously inhibited compression-induced apoptosis and ECM degradation of human NP cells. In the study described here, we found circRNA-CIDN's vital role during the pathophysiological process of IDD.
Accumulating studies demonstrate that ceRNA networks exert important functions in the initiation and development of various diseases [45,46]. Many classes of the ncRNA superfamily can act as ceRNAs, among which circRNAs as well as long noncoding RNAs (lncRNA) have presented key mechanistic roles by functioning as miRNA sponges [47], [48], [49]. It is worth noting that the kind of miRNA-sponge effects of circRNAs last longer than that of lncRNAs, as a result of its unique structure of covalently closed loop [50]. Using luciferase and RIP assays, we found that circRNA-CIDN inhibited compression-induced apoptosis and ECM degradation of human NP cells mainly through regulating miR-34a-5p. It has been reported that miR-34a-5p expression was significantly upregulated in NP tissues from IDD patients and that miR-34a-5p enhanced human NP cell apoptosis and extracellular matrix degradation [17,]. We further found that miR-34a-5p levels were dramatically up-regulated and exerted similar effects in human NP cells under compression. Subsequent assays were applied to screen the targets of miR-34a-5p and presented that a critical regulator of cell differentiation and proliferation, SIRT1 was validated to be a target of miR-34a-5p. SIRT1 is a nicotinamide-dependent class 3 histone deacetylase which regulates various cell fate decisions, and its deacetylase activity is significantly increased particularly under stresses [51,52]. More recently, evidence has indicated SIRT1 as a potential treatment agent for human degenerative IVD disease by restraining the apoptosis of NP cells [53,54]. Meanwhile, the activation of SIRT1 was reported to suppress cellular senescence, promote cell proliferation, accelerate autophagy, to maintain the NP extracellular matrix and keep the stability of the IVD [55], [56], [57]. Our results showed that SIRT1 expression was downregulated in compression treated human NP cells, which was remarkably rescued through inhibiting miR-34a-5p. We also demonstrated for the first time that SIRT1 overexpression induced by inhibiting miR-34a-5p exerted protective effects in IDD. Furthermore, in human IDD specimens, we observed a down-regulation of circRNA-CIDN and SIRT1 expression, and up-regulation of miR-34a-5p expression. We verified the positive correlation between circRNA-CIDN and SIRT1 mRNA expression, indicating that up-regulation of circRNA-CIDN could abrogate the repressive effect of miR-34a-5p on its target gene SIRT1. Thus, we proposed that the circRNA-CIDN/miR-34a-5p/SIRT1 axis served as a potential therapeutic target for treating IDD. For the analysis of expression levels of circRNA-CIDN, miR-34a-5p and SIRT1 mRNA and their correlation using patients tissue specimens, the IDD group were generally older than the scoliosis group. Since aging is a major etiological factor of IDD [58], the age difference might partially account for the present results and further investigation is needed. Still, the possibility of other critical genes as targets of circRNA-CIDN apart from SIRT1 that could exert important effects on IDD was not excluded.
As characterized in this study, circRNA-CIDN is generated by the back splicing of the exons 1 to 5 of the TRIM25 gene and mainly comprises the coding region of TRIM25, and the sequence of this circRNA is relatively conserved between human and rat. The target sites of miR-1271 in SIRT1 3′-UTR is also highly conserved in rat, which justify the application of ex vivo rat IVDs organ culture model. In our ex vivo compression-induced IDD model, we further validated the role of circRNA-CIDN in ameliorating IDD by acting as a miRNA sponge, as shown by the results of qRT-PCR assay, western blotting analysis, histological evaluation, immunohistochemical analysis and TUNEL apoptosis assessment. Taken together, the results strongly suggest that circRNA-CIDN is able to retard compression-induced apoptosis and ECM degradation of human NP cells by the miR-34a-5p/SIRT1 pathway. Therefore, targeting the circRNA-CIDN/miR-34a-5p/SIRT1 axis offers new therapeutic opportunities for treating IDD.
Declaration of Competing Interest
These authors have no conflict of interest to declare.
Acknowledgments
Funding sources
The funders had no role in study design, data collection, data analysis, interpretation or writing of the report. The corresponding author has full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81772391, 81974348) and the Fundamental Research Funds for the Central Universities (2017KFYXJJ248).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102679.
Contributor Information
Cao Yang, Email: caoyangunion@hust.edu.cn.
Yukun Zhang, Email: zhangyukuncom@126.com.
Appendix. Supplementary materials
References
- 1.Andersson G.B. Epidemiological features of chronic low-back pain. Lancet. 1999;354(9178):581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 2.Joud A., Petersson I.F., Englund M. Low back pain: epidemiology of consultations. Arthritis Care Res. 2012;64(7):1084–1088. doi: 10.1002/acr.21642. [DOI] [PubMed] [Google Scholar]
- 3.Froud R., Patterson S., Eldridge S., Seale C., Pincus T., Rajendran D. A systematic review and meta-synthesis of the impact of low back pain on people's lives. BMC Musculoskelet Disord. 2014;15:50. doi: 10.1186/1471-2474-15-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai D., Grad S. Advancing the cellular and molecular therapy for intervertebral disc disease. Adv Drug Deliv Rev. 2015;84:159–171. doi: 10.1016/j.addr.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Fontana G., See E., Pandit A. Current trends in biologics delivery to restore intervertebral disc anabolism. Adv Drug Deliv Rev. 2015;84:146–158. doi: 10.1016/j.addr.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Kepler C.K., Ponnappan R.K., Tannoury C.A., Risbud M.V., Anderson D.G. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13(3):318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Vo N.V., Hartman R.A., Yurube T., Jacobs L.J., Sowa G.A., Kang J.D. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 2013;13(3):331–341. doi: 10.1016/j.spinee.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachmeier B.E., Nerlich A., Mittermaier N., Weiler C., Lumenta C., Wuertz K. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur Spine J. 2009;18(11):1573. doi: 10.1007/s00586-009-1031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz A., Andersson G., Ortengren R., Haderspeck K., Nachemson A. Loads on the lumbar spine. Validation of a biomechanical analysis by measurements of intradiscal pressures and myoelectric signals. J Bone Joint Surg Am. 1982;64(5):713–720. [PubMed] [Google Scholar]
- 10.Adams M.A., McNally D.S., Dolan P. 'Stress' distributions inside intervertebral discs. The effects of age and degeneration. J Bone Joint Surg Br. 1996;78(6):965–972. doi: 10.1302/0301-620x78b6.1287. [DOI] [PubMed] [Google Scholar]
- 11.Wilke H.J., Neef P., Caimi M., Hoogland T., Claes L.E. New in vivo measurements of pressures in the intervertebral disc in daily life. Spine. 1999;24(8):755–762. doi: 10.1097/00007632-199904150-00005. [DOI] [PubMed] [Google Scholar]
- 12.Adams M.A., Roughley P.J. What is intervertebral disc degeneration, and what causes it? Spine. 2006;31(18):2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 13.Luoma K., Riihimaki H., Luukkonen R., Raininko R., Viikari-Juntura E., Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine. 2000;25(4):487–492. doi: 10.1097/00007632-200002150-00016. [DOI] [PubMed] [Google Scholar]
- 14.Yan Z., Pan Y., Wang S., Cheng M., Kong H., Sun C. Static compression induces ecm remodeling and integrin alpha2beta1 expression and signaling in a rat tail caudal intervertebral disc degeneration model. Spine. 2017;42(8):e448–ee58. doi: 10.1097/BRS.0000000000001856. [DOI] [PubMed] [Google Scholar]
- 15.Li S., Hua W., Wang K., Gao Y., Chen S., Liu W. Autophagy attenuates compression-induced apoptosis of human nucleus pulposus cells via MEK/ERK/NRF1/Atg7 signaling pathways during intervertebral disc degeneration. Exp Cell Res. 2018;370(1):87–97. doi: 10.1016/j.yexcr.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C.-.Q., Jiang L-S, Dai L-Y. Programmed cell death in intervertebral disc degeneration. Apoptosis. 2006;11(12):2079–2088. doi: 10.1007/s10495-006-0290-7. [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Zhang Y., Feng X., Li S., Gao Y., Wang K. Inhibition of microRNA-34a prevents IL-1beta-induced extracellular matrix degradation in nucleus pulposus by increasing GDF5 expression. Exp Biol Med. 2016;241(17):1924–1932. doi: 10.1177/1535370216657444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W.K., Yu X.H., Yang W., Wang C., He W.S., Yan Y.G. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(1):e12313. doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y., Zhang X., Li Z., Kong L., Huang Y. LncRNA hotair suppresses TNF-alpha induced apoptosis of nucleus pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed Pharmacother. 2018;107:729–737. doi: 10.1016/j.biopha.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Vicens Q., Westhof E. Biogenesis of circular RNAs. Cell. 2014;159(1):13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Shen S., Wu Y., Chen J., Xie Z., Huang K., Wang G. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis. 2019;78(6):826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and x linked inhibitor-of-apoptosis protein. Ann Rheum Dis. 2018;77(5):770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y., Li S., Geng W., Luo R., Liu W., Tu J. Sirtuin 3-dependent mitochondrial redox homeostasis protects against AGEs-induced intervertebral disc degeneration. Redox Biol. 2018;19:339–353. doi: 10.1016/j.redox.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reno C., Marchuk L., Sciore P., Frank C.B., Hart D.A. Rapid isolation of total RNA from small samples of hypocellular, dense connective tissues. Biotechniques. 1997;22(6):1082–1086. doi: 10.2144/97226bm16. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7(1):39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Mao H.J., Chen Q.X., Han B., Li F.C., Feng J., Shi Z.L. The effect of injection volume on disc degeneration in a rat tail model. Spine. 2011;36(16):E1062–E1069. doi: 10.1097/BRS.0b013e3182027d42. [DOI] [PubMed] [Google Scholar]
- 30.Ching C.T., Chow D.H., Yao F.Y., Holmes A.D. Changes in nuclear composition following cyclic compression of the intervertebral disc in an in vivo rat-tail model. Med Eng Phys. 2004;26(7):587–594. doi: 10.1016/j.medengphy.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 31.MacLean J.J., Lee C.R., Alini M., Iatridis J.C. The effects of short-term load duration on anabolic and catabolic gene expression in the rat tail intervertebral disc. J Orthop Res. 2005;23(5):1120–1127. doi: 10.1016/j.orthres.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Wu X., Liao Z., Wang K., Hua W., Liu X., Song Y. Targeting the IL-1β/IL-1Ra pathways for the aggregation of human islet amyloid polypeptide in an ex vivo organ culture system of the intervertebral disc. Exp Mol Med. 2019;51(9):110. doi: 10.1038/s12276-019-0310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartman R.A., Bell K.M., Debski R.E., Kang J.D., Sowa G.A. Novel ex-vivo mechanobiological intervertebral disc culture system. J Biomech. 2012;45(2):382–385. doi: 10.1016/j.jbiomech.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurakawa T., Kakutani K., Morita Y., Kato Y., Yurube T., Hirata H. Functional impact of integrin alpha5beta1 on the homeostasis of intervertebral discs: a study of mechanotransduction pathways using a novel dynamic loading organ culture system. Spine J. 2015;15(3):417–426. doi: 10.1016/j.spinee.2014.12.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junger S., Gantenbein-Ritter B., Lezuo P., Alini M., Ferguson S.J., Ito K. Effect of limited nutrition on in situ intervertebral disc cells under simulated-physiological loading. Spine. 2009;34(12):1264–1271. doi: 10.1097/BRS.0b013e3181a0193d. [DOI] [PubMed] [Google Scholar]
- 36.Lee C.R., Iatridis J.C., Poveda L., Alini M. In vitro organ culture of the bovine intervertebral disc: effects of vertebral endplate and potential for mechanobiology studies. Spine. 2006;31(5):515–522. doi: 10.1097/01.brs.0000201302.59050.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koerner J.D., Markova D.Z., Schroeder G.D., Rihn J.A., Hilibrand A.S., Vaccaro A.R. The effect of substance p on an intervertebral disc rat organ culture model. Spine. 2016;41(24):1851–1859. doi: 10.1097/BRS.0000000000001676. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 39.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K. Structure and degradation of circular RNAs regulate pkr activation in innate immunity. Cell. 2019;177(4):865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 40.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y. The landscape of circular rna in cancer. Cell. 2019;176(4):869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174(2):350–362. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Chen X., Xu D., Li S., Chan M.T.V., Wu W.K.K. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52(6):e12704. doi: 10.1111/cpr.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song J., Wang H.L., Song K.H., Ding Z.W., Wang H.L., Ma X.S. CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp Mol Med. 2018;50(8):94. doi: 10.1038/s12276-018-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Wang B., Zou M., Li J., Lu G., Zhang Q. CircSEMA4B targets miR-431 modulating IL-1beta-induced degradative changes in nucleus pulposus cells in intervertebral disc degeneration via WNT pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(11):3754–3768. doi: 10.1016/j.bbadis.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 45.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui X., Wang J., Guo Z., Li M., Li M., Liu S. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17(1):123. doi: 10.1186/s12943-018-0877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng Z., Yu C., Cui S., Wang H., Jin H., Wang C. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10(1):3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B., Wei W., Huang X., Xie X., Kong Y., Dai D. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song Y.X., Sun J.X., Zhao J.H., Yang Y.C., Shi J.X., Wu Z.H. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8(1):289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Kessler B. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 52.Han C., Gu Y., Shan H., Mi W., Sun J., Shi M. O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat Commun. 2017;8(1):1491. doi: 10.1038/s41467-017-01654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji M.L., Jiang H., Zhang X.J., Shi P.L., Li C., Wu H. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun. 2018;9(1):5051. doi: 10.1038/s41467-018-07360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z., Lin J., Nisar M., Chen T., Xu T., Zheng G. The sirt1/p53 axis in diabetic intervertebral disc degeneration pathogenesis and therapeutics. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/7959573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo J., Shao M., Lu F., Jiang J., Xia X. Role of sirt1 plays in nucleus pulposus cells and intervertebral disc degeneration. Spine. 2017;42(13):E757–Ee66. doi: 10.1097/BRS.0000000000001954. [DOI] [PubMed] [Google Scholar]
- 56.Jiang W., Zhang X., Hao J., Shen J., Fang J., Dong W. SIRT1 protects against apoptosis by promoting autophagy in degenerative human disc nucleus pulposus cells. Sci Rep. 2014;4:7456. doi: 10.1038/srep07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia X, Guo J, Lu F, Jiang J. SIRT1 plays a protective role in intervertebral disc degeneration in a puncture-induced rodent model. Spine. 2015;40(9):E515–24. [DOI] [PubMed]
- 58.Miller JA, Schmatz C, Schultz AB. Lumbar disc degeneration: correlation with age, sex, and spine level in 600 autopsy specimens. Spine. 1988;13(2):173–8. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.