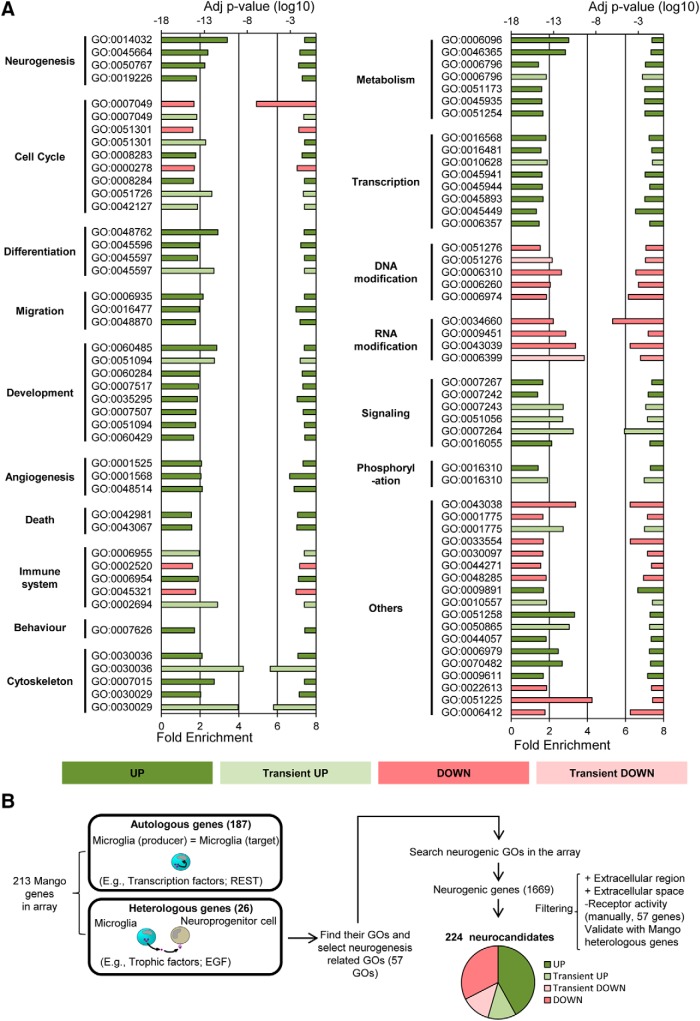
Figure 6.
Functional analysis of phagocytic microglia using DAVID and MANGO. A, Functional analysis of phagocytic microglia using DAVID software. Left axis represents the fold enrichment of each biological function and right axis represents the adjusted p value of each GO term. Key for the GO terms: GO:0014032, neural crest cell development; GO:0045664, regulation of neuron differentiation; GO:0050767, regulation of neurogenesis; GO:0019226, transmission of nerve impulse; GO:0007049, cell cycle; GO:0051301, cell division; GO:0008283, cell proliferation; GO:0000278, mitotic cell cycle; GO:0008284, positive regulation of cell proliferation; GO:0051726, regulation of cell cycle; GO:0042127, regulation of cell proliferation; GO:0048762, mesenchymal cell differentiation; GO:0045596, negative regulation of cell differentiation; GO:0045597, positive regulation of cell differentiation; GO:0006935, chemotaxis; GO:0016477, cell migration; GO:0048870, cell motility; GO:0060485, mesenchyme development; GO:0051094, positive regulation of developmental process; GO:0060284, regulation of cell development; GO:0007517, muscle organ development; GO:0035295, tube development; GO:0007507, heart development; GO:0051094, positive regulation of developmental process; GO:0060429, epithelium development; GO:0001525, angiogenesis; GO:0001568, blood vessel development; GO:0048514, blood vessel morphogenesis; GO:0042981, regulation of apoptosis; GO:0043067, regulation of programmed cell death; GO:0006955, immune response; GO:0002520, immune system development; GO:0006954, inflammatory response; GO:0045321, leukocyte activation; GO:0002694, regulation of leukocyte activation; GO:0007626, locomotory behavior; GO:0030036, actin cytoskeleton organization; GO:0007015, actin filament organization; GO:0030029, actin filament-based process; GO:0006096, glycolysis; GO:0046365, monosaccharide catabolic process; GO:0006796, phosphate metabolic process; GO:0006796, phosphate metabolic process; GO:0051173, positive regulation of nitrogen compound metabolic process; GO:0045935, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process; GO:0051254, positive regulation of RNA metabolic process; GO:0016568, chromatin modification; GO:0016481, negative regulation of transcription; GO:0010628, positive regulation of gene expression; GO:0045941, positive regulation of transcription; GO:0045944, positive regulation of transcription from RNA polymerase II promoter; GO:0045893, positive regulation of transcription, DNA-dependent; GO:0045449, regulation of transcription; GO:0006357, regulation of transcription from RNA polymerase II promoter; GO:0051276, chromosome organization; GO:0006310, DNA recombination; GO:0006260, DNA replication; GO:0006974, response to DNA damage stimulus; GO:0034660, ncRNA metabolic process; GO:0009451, RNA modification; GO:0043039, tRNA aminoacylation; GO:0006399 tRNA metabolic process; GO:0007267, cell–cell signaling; GO:0007242, intracellular signaling cascade; GO:0007243, protein kinase cascade; GO:0051056, regulation of small GTPase-mediated signal transduction; GO:0007264, small GTPase-mediated signal transduction; GO:0016055, Wnt receptor signaling pathway; GO:0016310, phosphorylation; GO:0043038, amino acid activation; GO:0001775, cell activation; GO:0033554, cellular response to stress; GO:0030097, hemopoiesis; GO:0044271, nitrogen compound biosynthetic process; GO:0048285, organelle fission; GO:0009891, positive regulation of biosynthetic process; GO:0010557, positive regulation of macromolecule biosynthetic process; GO:0051258, protein polymerization; GO:0050865, regulation of cell activation; GO:0044057, regulation of system process; GO:0006979, response to oxidative stress; GO:0070482, response to oxygen levels; GO:0009611, response to wounding; GO:0022613, ribonucleoprotein complex biogenesis; GO:0051225, spindle assembly; GO:0006412, translation. Left axis represents the fold enrichment of each biological function and right axis represents the adjusted p value of each GO term. Only statistically significant changes are shown. B, Diagram depicting the strategy followed to search for potential modulators of neurogenesis produced by phagocytic microglia in the arrays. The filtering started by differentiating the heterologous and autologous genes in the MANGO database. Then, GO terms related to neurogenesis were selected for the heterologous MANGO genes. Afterward, the molecules that presented the neurogenic GO terms were searched in the array. Finally, the candidate genes were filtered only to select those that appeared extracellularly (heterologous genes), and genes with receptor activity were manually discarded.