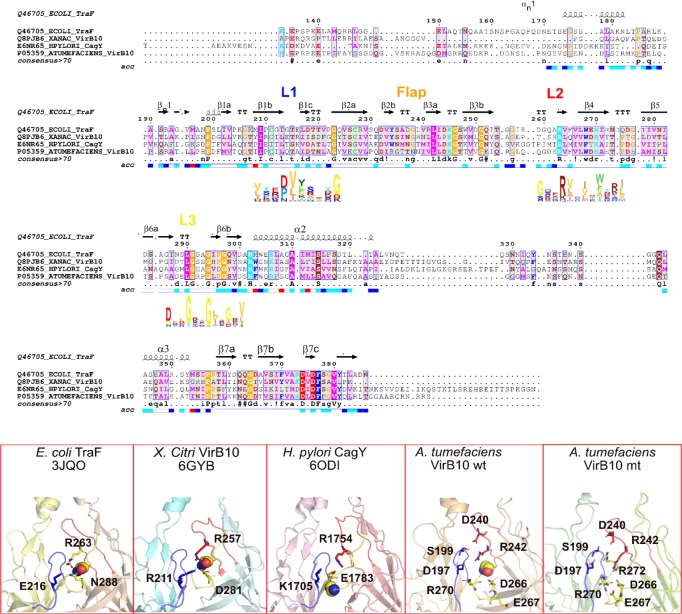
Figure 5.
Sequence alignment of VirB10CTD shown for the conjugation plasmid pKM101 T4S system (TraF), bacteria-killing effector translocator X. citri T4S system (VirB10), H. pylori Cag T4S system (CagY), and A. tumefaciens VirB10 using ENDscript/ESPript44 and HMM logo for loops L1, L2, and L3 displayed below the sequence using Skylign.45 The lower panel shows the conserved salt bridge triad in the inter-subunit interface for the above three systems and the corresponding salt bridge triad in the wild-type A. tumefaciens VirB10CTD oligomer (R242:D197:R270). Also shown for the wild type are other interacting residues involved in switching ionic interactions in the mutant G272R VirB10CTD. Loops L1 in blue, L2 in red, and L3 in yellow, and conserved G272 and corresponding middle Gly in the GxxGxxG motif in loop L3 are shown as spheres.