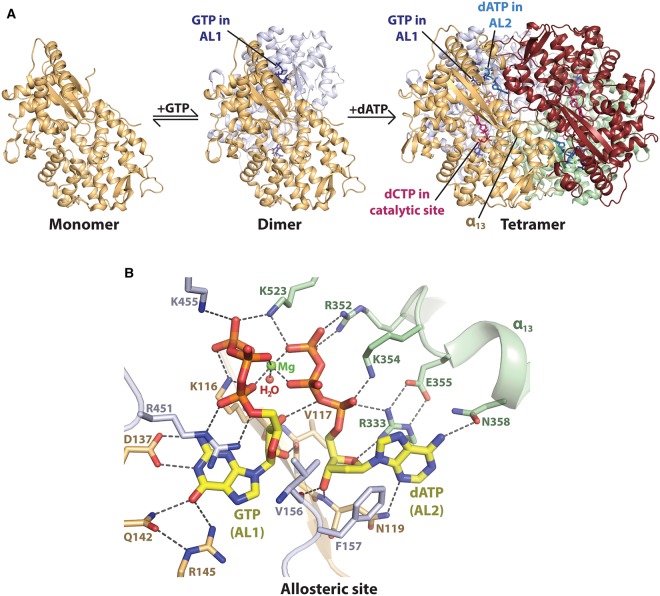
Figure 2. Nucleotide-dependent tetramerization of SAMHD1 HD domain.
(A) Ordered pathway for tetramer assembly, proposed by Hansen et al. [97], and elucidated structurally using X-ray crystallography [77,78,85–88,94,95,101]. The four protein monomers of SAMHD1 are shown in light orange, grey, maroon and light green. From left to right: The apo SAMHD1 HD domain is in a monomer–dimer equilibrium with GTP (dark blue), which stabilises dimerisation by coordinating AL1 (PDB code: 4RXO) [95]. dATP (light blue) co-ordinates AL2, and a magnesium ion (green sphere) bridges adjacent AL1–AL2 allosteric sites to stabilise the HD domain tetramer (PDB code: 4RXP) [95]. dCTP in the catalytic site is shown in pink. (B) GTP–dATP coordination at adjacent allosteric sites, AL1–AL2, of an HD domain tetramer (PDB code: 4TO0) [87]. GTP in AL1 and dATP in AL2 are shown in stick representation, with Cα atoms shown in yellow. Hydrogen-bonding interactions are represented by dashed lines.