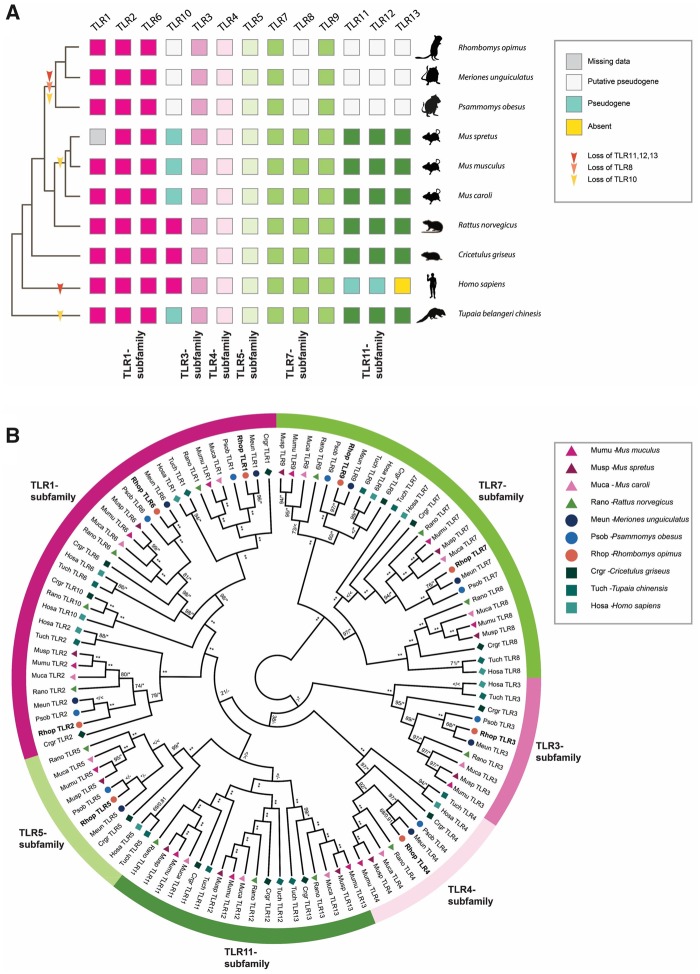
Fig. 2.
—TLR repertoire and ML phylogeny of investigated Gerbillinae, Rodentia, human, and Chinese tree shrew. (A) TLR repertoire of the investigated Gerbillinae, Rodentia, human, and Chinese tree shrew mapped onto a composite cladogram (see supplementary fig. S12, Supplementary Material online). The lineage-specific loss of TLR8 and all members of the TLR11-subfamily in Gerbillinae and other lineage-specific TLR losses are marked by arrows. Depicted in boxes colored by the six major subfamilies are the individual species’ TLR repertoires: TLR1-subfamily (dark pink), TLR3-subfamily (pink), TLR4-subfamily (light pink), TLR5-subfamily (light green), TLR7-subfamily (green), and TLR11-subfamily (dark green). Teal-colored boxes represent established pseudogenes, empty (white) boxes indicate putative pseudogenes, yellow boxes indicate complete absence of genes, and gray boxes represent missing information. (B) A maximum likelihood (ML) phylogeny of nucleotide sequences all full-length TLRs was created using RAxML with 100× topology and 500× bootstrap replicates. A MrBayes phylogeny with 20,000,000 generations and 25% burn-in was also created and the posterior probabilities added to the RAxML phylogeny. BS/PP; *, BS 100 or PP > 0.96; **, BS of 100 and PP>0.97; <, support values below 50/0.8; -, node not present in Bayesian analysis. Great gerbil genes are marked in bold and by orange circles. The six major TLR subfamilies are marked with colored bars and corresponding names. All investigated TLRs, including great gerbils, cluster well within each subfamily as well as being clearly separated into each TLR subfamily member.